Single-molecule protein sequencing with nanopores
IF 37.6
引用次数: 0
Abstract
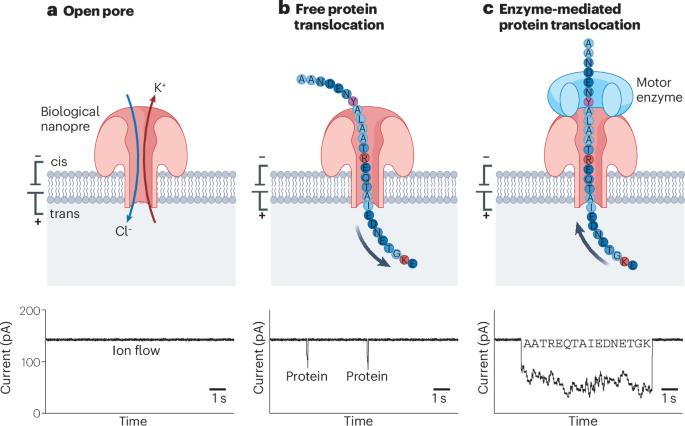
Protein sequencing and the identification of post-translational modifications are key to understanding cellular signalling pathways and metabolic processes in health and disease. Nanopores, that is, nanometre-sized holes in a membrane, were previously put to use for DNA and RNA sequencing, offering single-molecule sensitivity and long read lengths. This prompted efforts to engineer nanopores for the high-throughput sequencing of peptides and proteins. In this Review, we discuss research towards single-molecule protein sequencing using biological nanopores, investigating how their sensitivity allows the discrimination of all 20 amino acids. We outline how fingerprinting of proteins is facilitated by using motor proteins and electro-osmotic flow to promote the slow translocation of proteins through nanopores. Moreover, we examine applications of nanopores to the identification of post-translational modifications, highlighting the potential of this technology for fundamental and clinical proteomic studies. Finally, we outline the advantages and limitations of nanopore systems for protein sequencing and the challenges that remain to be overcome for realizing de novo nanopore protein sequencing. Biological nanopores can be designed for high-sensitivity sequencing of peptides and proteins. This Review discusses different approaches towards nanopore-based protein sequencing, outlining methods to distinguish amino acids, control protein translocation and detect post-translational modifications.
纳米孔单分子蛋白测序
蛋白质测序和鉴定翻译后修饰是理解健康和疾病中细胞信号通路和代谢过程的关键。纳米孔,即膜上纳米大小的孔,以前被用于DNA和RNA测序,提供单分子灵敏度和较长的读取长度。这促使人们努力设计纳米孔,用于多肽和蛋白质的高通量测序。在这篇综述中,我们讨论了利用生物纳米孔进行单分子蛋白质测序的研究,研究了它们的灵敏度如何允许所有20种氨基酸的区分。我们概述了如何利用马达蛋白和电渗透流来促进蛋白质通过纳米孔的缓慢易位,从而促进蛋白质的指纹识别。此外,我们研究了纳米孔在鉴定翻译后修饰方面的应用,强调了这项技术在基础和临床蛋白质组学研究中的潜力。最后,我们概述了纳米孔系统用于蛋白质测序的优势和局限性,以及实现从头纳米孔蛋白质测序仍需克服的挑战。生物纳米孔可用于多肽和蛋白质的高灵敏度测序。本文讨论了基于纳米孔的蛋白质测序的不同方法,概述了区分氨基酸、控制蛋白质易位和检测翻译后修饰的方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


