Integrated analysis of microRNA expression profiles in Takifugu obscurus kidney following Vibrio harveyi challenge
IF 2.4
3区 农林科学
Q1 FISHERIES
引用次数: 0
Abstract
MicroRNAs (miRNAs), a class of small non-coding RNAs critical for post-transcriptional gene regulation, serve as key immune modulators in vertebrates. However, their roles in teleost disease resistance remain underexplored. In the present study, a comprehensive miRNA transcriptome analysis was conducted in Takifugu obscurus under Vibrio harveyi challenge using high-throughput RNA sequencing. Three miRNA libraries (ToCG, ToEG1, and ToEG2) were constructed from kidney tissues of T. obscurus collected at 0 h (non-infected control), 6 h, and 24 h post-infection. Bioinformatics analysis identified 1093 miRNAs, including 195 novel miRNAs, and revealed 175 differentially expressed miRNAs (DEmiRNAs) across infection timepoints. Validation experiments via quantitative real-time PCR confirmed the expression patterns of 16 randomly selected DEmiRNAs, demonstrating high consistency with sequencing data. Functional enrichment analysis of DEmiRNAs highlighted significant associations with immune regulatory pathways, including mitogen-activated protein kinase signaling, Forkhead box O signaling, erythroblastic leukemia viral oncogene homolog signaling, and endocytosis pathways. Notably, several DEmiRNAs exhibited temporal expression patterns, suggesting stage-specific immunoregulatory functions during bacterial pathogenesis. This study suggests a potential miRNA regulatory network in T. obscurus during V. harveyi infection, providing crucial insights into conserved and species-specific miRNA-mediated immune mechanisms in teleosts.
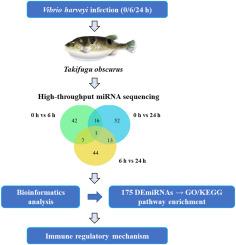
哈维氏弧菌侵染后暗鲀肾脏microRNA表达谱的综合分析
MicroRNAs (miRNAs)是一类对转录后基因调控至关重要的小非编码rna,是脊椎动物的关键免疫调节剂。然而,它们在硬骨鱼疾病抗性中的作用仍未得到充分探索。在本研究中,利用高通量RNA测序技术,对哈维氏弧菌侵染下的暗鲀进行了全面的miRNA转录组分析。从感染后0 h(未感染对照)、6 h和24 h收集的隐壁绦虫肾脏组织中构建了三个miRNA文库(ToCG、ToEG1和ToEG2)。生物信息学分析鉴定了1093种mirna,其中包括195种新型mirna,并揭示了175种不同感染时间点的差异表达mirna (demirna)。通过实时荧光定量PCR验证实验,证实了随机选取的16个demirna的表达模式,与测序数据具有较高的一致性。DEmiRNAs的功能富集分析强调了与免疫调节途径的显著关联,包括丝裂原激活的蛋白激酶信号、叉头盒O信号、红母细胞白血病病毒癌基因同源信号和内吞作用途径。值得注意的是,一些demirna表现出时间表达模式,表明在细菌发病过程中具有特定阶段的免疫调节功能。这项研究表明,在哈维伊梭菌感染期间,暗箱鱼体内存在潜在的miRNA调控网络,为硬鱼中保守的和物种特异性的miRNA介导的免疫机制提供了重要的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
6.20
自引率
6.90%
发文量
206
审稿时长
49 days
期刊介绍:
Developmental and Comparative Immunology (DCI) is an international journal that publishes articles describing original research in all areas of immunology, including comparative aspects of immunity and the evolution and development of the immune system. Manuscripts describing studies of immune systems in both vertebrates and invertebrates are welcome. All levels of immunological investigations are appropriate: organismal, cellular, biochemical and molecular genetics, extending to such fields as aging of the immune system, interaction between the immune and neuroendocrine system and intestinal immunity.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


