Deep learning-based cross-device standardization of surface-enhanced Raman spectroscopy for enhanced bacterial recognition
IF 4.6
2区 化学
Q1 SPECTROSCOPY
Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy
Pub Date : 2025-09-19
DOI:10.1016/j.saa.2025.126931
引用次数: 0
Abstract
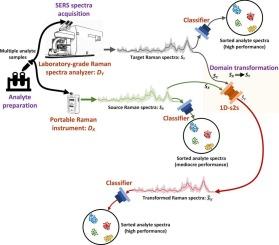
Surface-enhanced Raman spectroscopy (SERS) is a powerful, label-free technique for pathogen detection; however, its broader adoption in clinical diagnostics is hindered by inconsistent spectral quality across portable and laboratory-grade instruments, limited cross-device reproducibility, and the poor generalizability of existing machine learning approaches. These limitations restrict reliable and rapid pathogen identification at the point of care. To address this gap, we collected SERS spectra from analytes spread on silver nanorod (AgNR) substrates using four portable Raman systems (Enwave, Tec5, First Defender, and Rapid ID) and one laboratory-grade reference device (Renishaw). The dataset included 20 analyte classes representing clinically relevant bacterial signatures and reference compounds. We propose a deep learning framework comprising: (1) SERS-D2DNet, a one-dimensional sequence-to-sequence neural network that transforms spectra from portable devices into high-fidelity laboratory-grade equivalents, and (2) SuperRaman, a lightweight super-operational neural network (Super-ONN) for efficient multiclass bacterial classification. Primary and ablation studies confirm the complementary role of domain transformation and classification, demonstrating improved feature separability and reduced misclassification rates. Quantitative results show that SERS-D2DNet reduced mean absolute error to 0.01 and increased R2 to over 98 % across devices, while SuperRaman achieved up to 100 % classification accuracy post-transformation. Compared to existing approaches, SERS-D2DNet delivered the lowest MAE (0.024 to 0.034), while SuperRaman surpassed state-of-the-art classifiers. The combined framework requires only 6.6 million parameters, a compact 9 MB footprint, and a 3.27 ms inference time, making it well-suited for portable deployment. This study establishes a scalable, real-time solution for rapid sepsis detection and pathogen identification, bridging the performance gap between portable and laboratory-grade SERS systems.
基于深度学习的表面增强拉曼光谱跨设备标准化,以增强细菌识别
表面增强拉曼光谱(SERS)是一种强大的、无标签的病原体检测技术;然而,它在临床诊断中的广泛采用受到便携式和实验室级仪器之间不一致的光谱质量,有限的跨设备可重复性以及现有机器学习方法的不良通用性的阻碍。这些限制限制了在护理点进行可靠和快速的病原体鉴定。为了解决这一空白,我们使用四个便携式拉曼系统(Enwave, Tec5, First Defender和Rapid ID)和一个实验室级参考设备(Renishaw)收集了分布在银纳米棒(AgNR)基片上的分析物的SERS光谱。该数据集包括20个分析物类别,代表临床相关的细菌特征和参考化合物。我们提出了一个深度学习框架,包括:(1)SERS-D2DNet,一个将便携式设备的光谱转换为高保真实验室级等效光谱的一维序列对序列神经网络;(2)SuperRaman,一个用于高效多类细菌分类的轻量级超操作神经网络(Super-ONN)。初步研究和消融研究证实了域变换和分类的互补作用,证明了特征可分离性的提高和误分类率的降低。定量结果表明,SERS-D2DNet将设备间的平均绝对误差降低到0.01,将R2提高到98%以上,而SuperRaman在转换后的分类准确率高达100%。与现有方法相比,SERS-D2DNet提供了最低的MAE(0.024至0.034),而SuperRaman超越了最先进的分类器。合并后的框架只需要660万个参数,紧凑的9 MB内存和3.27 ms推理时间,使其非常适合便携式部署。本研究建立了一种可扩展的实时解决方案,用于快速败血症检测和病原体鉴定,弥合了便携式和实验室级SERS系统之间的性能差距。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.40
自引率
11.40%
发文量
1364
审稿时长
40 days
期刊介绍:
Spectrochimica Acta, Part A: Molecular and Biomolecular Spectroscopy (SAA) is an interdisciplinary journal which spans from basic to applied aspects of optical spectroscopy in chemistry, medicine, biology, and materials science.
The journal publishes original scientific papers that feature high-quality spectroscopic data and analysis. From the broad range of optical spectroscopies, the emphasis is on electronic, vibrational or rotational spectra of molecules, rather than on spectroscopy based on magnetic moments.
Criteria for publication in SAA are novelty, uniqueness, and outstanding quality. Routine applications of spectroscopic techniques and computational methods are not appropriate.
Topics of particular interest of Spectrochimica Acta Part A include, but are not limited to:
Spectroscopy and dynamics of bioanalytical, biomedical, environmental, and atmospheric sciences,
Novel experimental techniques or instrumentation for molecular spectroscopy,
Novel theoretical and computational methods,
Novel applications in photochemistry and photobiology,
Novel interpretational approaches as well as advances in data analysis based on electronic or vibrational spectroscopy.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


