Addition to “Structure-Based Ligand Discovery Targeting the Transmembrane Domain of Frizzled Receptor FZD7”
IF 6.8
1区 医学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
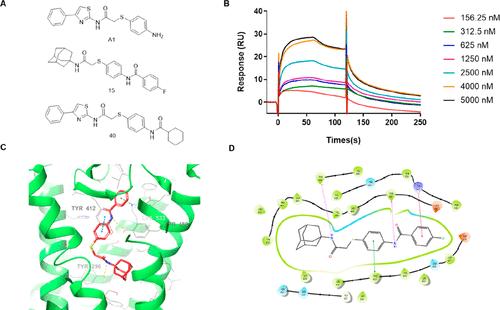
Figure A1. Evaluation of Compound 15 binding to FZD7-TMD. Chemical structures of intermediate A1, compound 15, and compound 40 (A). SPR analysis of the interaction between compound 15 and FZD7-TMD (B). Molecular docking of compound 15 with FZD7 (C) and its interactions with the FZD7 binding pocket (D). Figure A2. Binding analysis of compound F7H and compound 28 using MST assay. Dose–response binding curves of compound F7H (A), compound 28 (B) with FZD7-TMD protein. Figure A3. Thermal shift assay to assess compound 28 binding to various proteins. FZD1 (A), FZD4 (B), FZD6 (C), FZD7 (D), GPR15 (E), and compounds only (F). Figure A4. TopFlash assay in FZD1–10 knockout cells. Cells were transfected with PTT5 (A), WT-FZD7 (B), or the mutant K533A (C) and then treated with IWP2 or compound 28. Figure A5. Molecular docking results of compound 28 with firefly luciferase (PDB: 1LCI). This article references 4 other publications. This article has not yet been cited by other publications.
补充“针对卷曲受体FZD7跨膜结构域的基于结构的配体发现”
图A1。化合物15与FZD7-TMD结合的评价。中间体A1、化合物15和化合物40 (A)的化学结构。化合物15与FZD7-TMD相互作用的SPR分析(B)。化合物15与FZD7的分子对接(C)及其与FZD7结合袋的相互作用(D)。图A2。用MST法分析化合物F7H和化合物28的结合。化合物F7H (A)、化合物28 (B)与FZD7-TMD蛋白的剂量-反应结合曲线。图A3。热移试验评估化合物28与各种蛋白质的结合。FZD1 (A), FZD4 (B), FZD6 (C), FZD7 (D), GPR15 (E)和仅化合物(F)。图A4。FZD1-10敲除细胞的TopFlash检测。用PTT5 (A)、WT-FZD7 (B)或突变体K533A (C)转染细胞,然后用IWP2或化合物28处理。图A5。化合物28与萤火虫荧光素酶(PDB: 1LCI)的分子对接结果。本文引用了其他4篇出版物。这篇文章尚未被其他出版物引用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Medicinal Chemistry
医学-医药化学
CiteScore
4.00
自引率
11.00%
发文量
804
审稿时长
1.9 months
期刊介绍:
The Journal of Medicinal Chemistry is a prestigious biweekly peer-reviewed publication that focuses on the multifaceted field of medicinal chemistry. Since its inception in 1959 as the Journal of Medicinal and Pharmaceutical Chemistry, it has evolved to become a cornerstone in the dissemination of research findings related to the design, synthesis, and development of therapeutic agents.
The Journal of Medicinal Chemistry is recognized for its significant impact in the scientific community, as evidenced by its 2022 impact factor of 7.3. This metric reflects the journal's influence and the importance of its content in shaping the future of drug discovery and development. The journal serves as a vital resource for chemists, pharmacologists, and other researchers interested in the molecular mechanisms of drug action and the optimization of therapeutic compounds.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


