NovumRNA: Accurate prediction of non-canonical tumor antigens from RNA sequencing data
IF 4.1
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
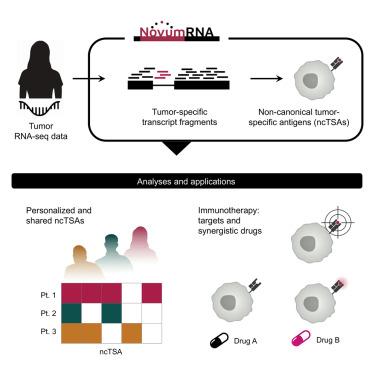
Non-canonical tumor-specific antigens (ncTSAs) can expand the pool of targets for cancer immunotherapy, but require robust and comprehensive computational pipelines for their prediction. Here, we present NovumRNA, a fully automated Nextflow pipeline for predicting different classes of ncTSAs from patients’ RNA sequencing data. We extensively benchmarked NovumRNA using publicly available and newly generated datasets, demonstrating the robustness of its analytical modules and predictions. NovumRNA analysis of RNA-seq data from colorectal cancer organoids and patients’ tumor samples revealed comparable ncTSA potential for microsatellite stable and unstable tumors and candidate therapeutic targets for patients with low tumor mutational burden. Finally, our investigation of glioblastoma cell lines demonstrated increased ncTSAs burden upon indisulam treatment, and detection by NovumRNA of therapy-induced ncTSAs, which we could validate experimentally. These findings underscore the potential of NovumRNA for identifying synergistic drugs and novel therapeutic targets for immunotherapy, which could ultimately extend its benefit to a broader patient population.
NovumRNA:从RNA测序数据中准确预测非典型肿瘤抗原
非典型肿瘤特异性抗原(ncTSAs)可以扩大癌症免疫治疗的靶标池,但需要强大而全面的计算管道来预测它们。在这里,我们提出了NovumRNA,这是一种全自动Nextflow管道,用于从患者的RNA测序数据中预测不同类型的ncTSAs。我们使用公开可用的和新生成的数据集对NovumRNA进行了广泛的基准测试,证明了其分析模块和预测的稳健性。来自结直肠癌类器官和患者肿瘤样本的RNA-seq数据的NovumRNA分析显示,ncTSA在微卫星稳定和不稳定肿瘤以及低肿瘤突变负担患者的候选治疗靶点方面具有相当的潜力。最后,我们对胶质母细胞瘤细胞系的研究表明,胰岛素治疗后ncTSAs负担增加,NovumRNA检测到治疗诱导的ncTSAs,我们可以通过实验验证。这些发现强调了NovumRNA在确定协同药物和新的免疫治疗靶点方面的潜力,这可能最终将其益处扩展到更广泛的患者群体。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


