MicroDFBEST: A dCas12b-derived dual-function base editor with programmable editing characteristics for microbial genetic engineering
IF 4.4
2区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Base editors (BEs) enable precise genome editing, but their use in microbes remains limited by restricted mutagenesis capabilities and narrow editing windows. Here, we reported MicroDFBEST, a novel dual-function base editor (DFBE) for microbes, by fusing the high-activity deaminases evoCDA1 and TadA9 with nuclease-deficient Cas12b from Bacillus hisashii (dBhCas12b). This engineered system enables simultaneous C-to-T and A-to-G editing within a 26–33 nt window, the broadest range reported for microbial DFBEs. The editing characteristics of MicroDFBEST can be easily adjusted by changing fusion protein expression and editing generations to create diverse mutant libraries. We show that the MicroDFBEST system enables both flexible gene expression modulation via random promoter (PylbP) diversification and targeted protein evolution through mutational hotspot scanning in native genomic contexts. This study offers a versatile platform enabling in situ gene regulation (e.g., biosynthetic gene clusters activation) and protein evolution (e.g., chassis optimization), with broad synthetic biology utility.
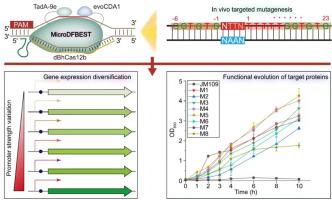
MicroDFBEST:一种dcas12b衍生的双功能碱基编辑器,具有可编程编辑特性,用于微生物基因工程
碱基编辑器(BEs)能够实现精确的基因组编辑,但它们在微生物中的应用仍然受到有限的诱变能力和狭窄的编辑窗口的限制。在这里,我们报道了MicroDFBEST,一种新的微生物双功能碱基编辑器(DFBE),通过将高活性脱氨酶evoCDA1和TadA9与来自hisashii芽孢杆菌的核酸酶缺陷Cas12b (dBhCas12b)融合。该工程系统能够在26 - 33nt的窗口内同时进行C-to-T和a -to- g编辑,这是目前报道的微生物DFBEs中范围最广的。通过改变融合蛋白表达和编辑代数,可以轻松调整MicroDFBEST的编辑特性,从而创建不同的突变文库。我们发现MicroDFBEST系统既可以通过随机启动子(PylbP)多样化灵活地调节基因表达,也可以通过突变热点扫描在原生基因组环境中实现靶向蛋白进化。该研究为原位基因调控(如生物合成基因簇激活)和蛋白质进化(如底盘优化)提供了一个多功能平台,具有广泛的合成生物学实用性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Synthetic and Systems Biotechnology
BIOTECHNOLOGY & APPLIED MICROBIOLOGY-
CiteScore
6.90
自引率
12.50%
发文量
90
审稿时长
67 days
期刊介绍:
Synthetic and Systems Biotechnology aims to promote the communication of original research in synthetic and systems biology, with strong emphasis on applications towards biotechnology. This journal is a quarterly peer-reviewed journal led by Editor-in-Chief Lixin Zhang. The journal publishes high-quality research; focusing on integrative approaches to enable the understanding and design of biological systems, and research to develop the application of systems and synthetic biology to natural systems. This journal will publish Articles, Short notes, Methods, Mini Reviews, Commentary and Conference reviews.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


