Epigenetic imprinting in innate lymphoid cell precursors directs the lineage segregation of innate lymphoid cells
IF 27.6
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
Abstract
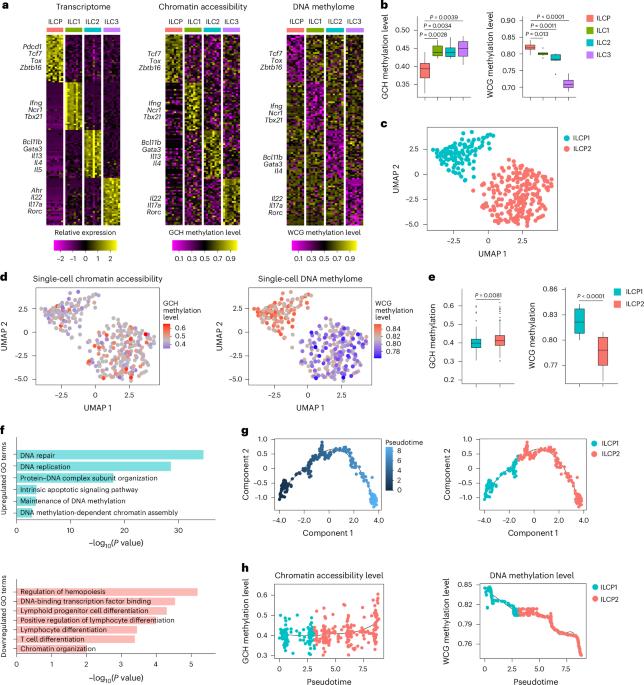
Innate lymphoid cells (ILCs) are essential for mucosal homeostasis, but the epigenetic regulation of their lineage segregation remains elusive. Here we simultaneously profiled the single-cell DNA methylome, chromatin accessibility and transcriptome of ILC subsets and ILC precursors (ILCPs) and found that ILCPs could be divided into two subgroups (ILCP1 and ILCP2). ILCP2s had highly heterogeneous DNA methylation profiles and could be divided into three groups according to their DNA methylation characteristics, which matched those of ILC subsets. We identified the signature methylation regions (SMRs) of each ILC subset and traced the DNA methylation imprinting during ILCP differentiation. ILCP2s with hypomethylated SMRs characteristic of ILC subsets differentiated into those subsets. DNA methylation editing of SMRs suppressed ILC lineage segregation, while deletion of Dnmt1 in ILCPs abrogated the heterogeneous distribution of SMRs and resulted in ILC differentiation defects. These findings provide evidence that epigenetic imprinting determines lineage segregation during immune cell development. Wang and colleagues show that specific DNA methylation profiles mark the ILC progenitors (ILCP) that would differentiate into ILC1, ILC2 or ILC3 subsets.
先天淋巴样细胞前体的表观遗传印迹指导先天淋巴样细胞的谱系分离。
先天淋巴样细胞(ILCs)对粘膜稳态至关重要,但其谱系分离的表观遗传调控仍然难以捉摸。在这里,我们同时分析了ILC亚群和ILC前体(ILCPs)的单细胞DNA甲基组、染色质可及性和转录组,发现ILCPs可以分为两个亚群(ILCP1和ILCP2)。ILCP2s具有高度异质性的DNA甲基化谱,根据其DNA甲基化特征可分为三组,这与ILC亚群的DNA甲基化特征相匹配。我们鉴定了每个ILC子集的特征甲基化区域(SMRs),并追踪了ILCP分化过程中的DNA甲基化印记。具有ILC亚群低甲基化SMRs特征的ILCP2s分化为这些亚群。SMRs的DNA甲基化编辑抑制了ILC谱系分离,而在ilcp中删除Dnmt1则消除了SMRs的异质分布,导致ILC分化缺陷。这些发现为表观遗传印迹决定免疫细胞发育过程中的谱系分离提供了证据。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


