Rapid learning of neural circuitry from holographic ensemble stimulation enabled by model-based compressed sensing
IF 20
1区 医学
Q1 NEUROSCIENCES
引用次数: 0
Abstract
Discovering how computations are implemented in the brain at the level of monosynaptic connectivity requires probing for connections from potentially thousands of presynaptic candidate neurons. Two-photon optogenetics is a promising technology for mapping such connectivity via sequential stimulation of individual neurons while recording postsynaptic responses intracellularly. However, this technique is currently not scalable because stimulating neurons one by one requires prohibitively long experiments. Here we developed novel computational tools that, when combined, enable learning of monosynaptic connectivity from high-speed holographic ensemble stimulation. First, we developed a model-based compressed sensing algorithm that identifies connections from postsynaptic responses evoked by stimulating many neurons at once, greatly increasing mapping efficiency. Second, we developed a deep-learning method that isolates the postsynaptic response to each stimulus, allowing stimulation to rapidly switch between ensembles without waiting for the postsynaptic response to return to baseline. Together, our system increases the throughput of connectivity mapping by an order of magnitude, facilitating discovery of the circuitry underlying neural computations. The authors develop a new computational system for high-throughput mapping of synaptic connectivity using two-photon holographic optogenetics and intracellular recordings.
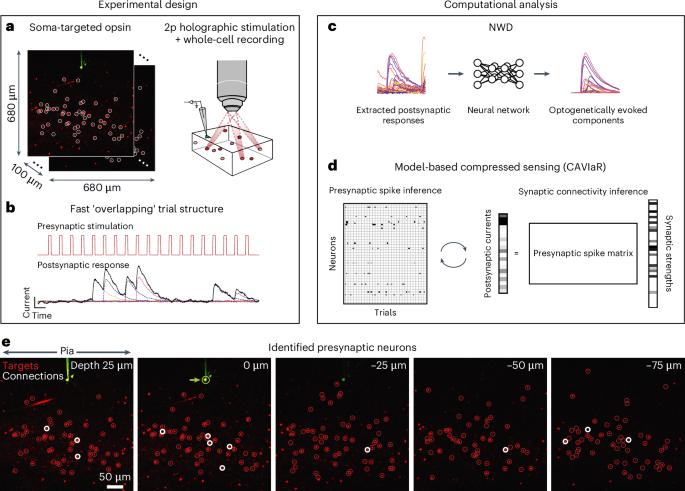
基于模型压缩感知的全息集合刺激下神经回路的快速学习。
在单突触连接水平上发现计算是如何在大脑中实现的,需要探测潜在的数千个突触前候选神经元的连接。双光子光遗传学是一种很有前途的技术,可以通过对单个神经元的顺序刺激来绘制这种连接,同时记录细胞内的突触后反应。然而,这种技术目前还不能扩展,因为逐个刺激神经元需要非常长的实验时间。在这里,我们开发了新的计算工具,当结合起来时,可以从高速全息集合刺激中学习单突触连接。首先,我们开发了一种基于模型的压缩感知算法,该算法可以从同时刺激多个神经元引起的突触后反应中识别连接,从而大大提高了映射效率。其次,我们开发了一种深度学习方法,分离每个刺激的突触后反应,允许刺激在集合之间快速切换,而无需等待突触后反应返回基线。总之,我们的系统将连接映射的吞吐量提高了一个数量级,有助于发现神经计算背后的电路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature neuroscience
医学-神经科学
CiteScore
38.60
自引率
1.20%
发文量
212
审稿时长
1 months
期刊介绍:
Nature Neuroscience, a multidisciplinary journal, publishes papers of the utmost quality and significance across all realms of neuroscience. The editors welcome contributions spanning molecular, cellular, systems, and cognitive neuroscience, along with psychophysics, computational modeling, and nervous system disorders. While no area is off-limits, studies offering fundamental insights into nervous system function receive priority.
The journal offers high visibility to both readers and authors, fostering interdisciplinary communication and accessibility to a broad audience. It maintains high standards of copy editing and production, rigorous peer review, rapid publication, and operates independently from academic societies and other vested interests.
In addition to primary research, Nature Neuroscience features news and views, reviews, editorials, commentaries, perspectives, book reviews, and correspondence, aiming to serve as the voice of the global neuroscience community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


