From phylogeny to photonics: A smart spectroscopic approach assisted with neural networks for rapid typing of Enterobacteriaceae isolates from chicken meat
IF 3.5
3区 医学
Q3 IMMUNOLOGY
引用次数: 0
Abstract
Bacteria are ubiquitous organisms; while many are harmless, some can cause serious disease outbreaks. Nearly a quarter of the global food supply is ruined by microbial activity, such as spoilage microorganisms or disease-causing pathogens, leading to substantial economic losses and human safety problems. Most mishaps associated with bacterial contamination could be avoided if a 100 % accurate, easy-to-use, rapid detector/identifier of bacteria was available. Compared to the time-consuming and complicated conventional molecular characterization methods, laser-induced breakdown spectroscopy (LIBS) combined with appropriate chemometric analysis provides a potential approach for the rapid and reliable characterization of bacterial species. This study identified isolated bacterial contaminants from store-bought chicken fillets using molecular characterization of the polymerase chain reaction (PCR) products of small subunit 16S rRNA sequences. Eight different strains were identified, all belonging to the family Enterobacteriaceae. Phylogenetic relationships were investigated using a constructed phylogenetic tree. In this study, nanoparticle-enhanced laser-induced breakdown spectroscopy (NELIBS) was used to differentiate bacterial samples based on their elemental composition. Spectral data were analyzed using principal component analysis (PCA) to explore the variance between the pure bacterial samples. With a cumulative variance between 95 % and 97 %, sample groups at closely related taxonomic levels (genus, species, and subspecies) could be discriminated. The robustness of the LIBS characterization was tested in the presence of a second bacterial species in the ablated specimen using mixtures of bacterial cultures at different concentrations. Using artificial neural networks (ANN), the spectral data were classified with an accuracy of up to 100 %.
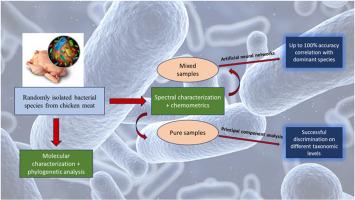
从系统发育到光子学:一种智能光谱方法辅助神经网络快速分型鸡肉肠杆菌科分离物。
细菌是无处不在的有机体;虽然许多病毒是无害的,但有些病毒会导致严重的疾病爆发。全球近四分之一的粮食供应被微生物活动破坏,如腐败微生物或致病病原体,导致重大经济损失和人体安全问题。如果有100%准确、易于使用、快速的细菌检测器/标识符,大多数与细菌污染有关的事故都可以避免。与耗时且复杂的传统分子表征方法相比,激光诱导击穿光谱(LIBS)与适当的化学计量学分析相结合,为快速、可靠地表征细菌种类提供了一种潜在的方法。本研究利用小亚基16S rRNA序列的聚合酶链反应(PCR)产物的分子特征鉴定了从商店购买的鸡柳中分离的细菌污染物。鉴定出8株不同的菌株,均属于肠杆菌科。利用构建的系统发育树研究系统发育关系。在这项研究中,纳米粒子增强激光诱导击穿光谱(NELIBS)用于区分细菌样品的元素组成。采用主成分分析(PCA)对光谱数据进行分析,探讨纯细菌样品之间的差异。累积方差在95% ~ 97%之间,可以在相近的分类水平(属、种、亚种)上区分样本群。使用不同浓度的细菌培养混合物,在消融标本中存在第二种细菌物种的情况下,测试了LIBS表征的稳健性。利用人工神经网络(ANN)对光谱数据进行分类,准确率高达100%。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Microbial pathogenesis
医学-免疫学
CiteScore
7.40
自引率
2.60%
发文量
472
审稿时长
56 days
期刊介绍:
Microbial Pathogenesis publishes original contributions and reviews about the molecular and cellular mechanisms of infectious diseases. It covers microbiology, host-pathogen interaction and immunology related to infectious agents, including bacteria, fungi, viruses and protozoa. It also accepts papers in the field of clinical microbiology, with the exception of case reports.
Research Areas Include:
-Pathogenesis
-Virulence factors
-Host susceptibility or resistance
-Immune mechanisms
-Identification, cloning and sequencing of relevant genes
-Genetic studies
-Viruses, prokaryotic organisms and protozoa
-Microbiota
-Systems biology related to infectious diseases
-Targets for vaccine design (pre-clinical studies)
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


