Treatment-agnostic joint modeling of longitudinal circulating tumor dna predicts survival across first-line regimens in metastatic non-squamous NSCLC
IF 4.7
3区 医学
Q1 PHARMACOLOGY & PHARMACY
引用次数: 0
Abstract
Background and Purpose
To develop a treatment-agnostic joint model leveraging longitudinal circulating tumor DNA (ctDNA) dynamics to predict overall survival (OS) across different treatment regimens without incorporating treatment information using data from the IMpower150 trial in treatment-naïve metastatic non-squamous NSCLC patients.
Experimental approach
Patients were randomized 1:1:1 to receive ABCP, ACP, or BCP, with plasma samples collected at baseline and multiple time points to track longitudinal ctDNA changes. The joint model followed a two-step approach by combining two separate submodels: a disease submodel for ctDNA dynamics and a survival submodel for time-to-event analysis. Random effects are derived from the nonlinear mixed effects model for ctDNA dynamics. In the subsequent step, these individual random effects are incorporated as covariates in the survival submodel. A landmark modeling approach was used where ctDNA data from the first 21 weeks posttreatment was used to predicts OS beyond 21 weeks.
Key Results
Among 466 participants, 348 had detectable ctDNA at one or more time points and were stratified into training (n = 181) and test (n = 167) sets. Fourteen ctDNA summary metrics were assessed, with median allele frequency for known/likely mutations emerging as the top-performing metric. Overall, the treatment-agnostic model's predicted survival curves closely matched the observed ones across treatment arms in both training and test sets. Predicted median OS and 2-/3-year OS rates aligned well for ABCP, ACP, and BCP, with discrepancies generally under 20 %. Although the model tended to overpredict OS for ACP—likely due to a small sample size—the final IMpower150 analysis reported median OS values within 10 % of our predictions.
Conclusions & Implications
Early ctDNA monitoring and modeling have the potential to significantly enhance treatment personalization. By enabling the early prediction of patient outcomes across various treatment regimens, this treatment-agnostic model supports more informed clinical decision-making, ultimately improving patient management and outcomes in oncology.
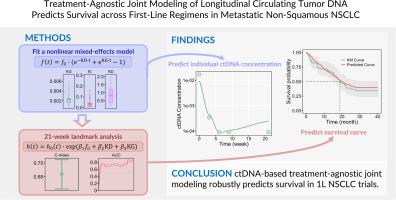
纵向循环肿瘤DNA联合建模预测转移性非鳞状非小细胞肺癌一线治疗方案的生存率。
背景和目的:利用纵向循环肿瘤DNA (ctDNA)动力学来预测不同治疗方案的总生存期(OS),而不纳入IMpower150试验中treatment-naïve转移性非鳞状NSCLC患者的治疗信息。实验方法:患者以1:1:1的比例随机接受ABCP、ACP或BCP,并在基线和多个时间点收集血浆样本,以跟踪纵向ctDNA变化。联合模型采用两步方法,结合了两个独立的子模型:ctDNA动力学的疾病子模型和事件时间分析的生存子模型。随机效应是由ctDNA动力学的非线性混合效应模型导出的。在随后的步骤中,这些个体随机效应作为协变量纳入生存子模型。采用里程碑式建模方法,使用治疗后前21周的ctDNA数据预测21周后的OS。关键结果:在466名参与者中,348名在一个或多个时间点检测到ctDNA,并分为训练组(n=181)和测试组(n=167)。评估了14个ctDNA汇总指标,已知/可能突变的中位等位基因频率成为表现最好的指标。总的来说,治疗不可知模型的预测生存曲线在训练集和测试集中与观察到的治疗组的生存曲线非常吻合。ABCP、ACP和BCP的预测中位OS和2 /3年OS率一致,差异一般在20%以下。尽管该模型倾向于高估acp的OS(可能是由于样本量小),但最终的IMpower150分析报告的OS中值在我们预测的10%以内。结论和意义:早期ctDNA监测和建模具有显著提高治疗个性化的潜力。通过早期预测不同治疗方案的患者预后,这种治疗不可知模型支持更明智的临床决策,最终改善肿瘤患者管理和预后。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.60
自引率
2.20%
发文量
248
审稿时长
50 days
期刊介绍:
The journal publishes research articles, review articles and scientific commentaries on all aspects of the pharmaceutical sciences with emphasis on conceptual novelty and scientific quality. The Editors welcome articles in this multidisciplinary field, with a focus on topics relevant for drug discovery and development.
More specifically, the Journal publishes reports on medicinal chemistry, pharmacology, drug absorption and metabolism, pharmacokinetics and pharmacodynamics, pharmaceutical and biomedical analysis, drug delivery (including gene delivery), drug targeting, pharmaceutical technology, pharmaceutical biotechnology and clinical drug evaluation. The journal will typically not give priority to manuscripts focusing primarily on organic synthesis, natural products, adaptation of analytical approaches, or discussions pertaining to drug policy making.
Scientific commentaries and review articles are generally by invitation only or by consent of the Editors. Proceedings of scientific meetings may be published as special issues or supplements to the Journal.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


