qPCR-based method to measure plasmid internalization efficiency of non-viral nanoparticle carriers in adherent cell model
IF 2.5
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Non-viral nanoparticle-mediated transfection systems have gained wide acceptance in modern pharmaceutical and biotechnological applications, including RNA delivery for COVID-19 vaccines. However, these systems may show low transfection and transcription efficiencies, potentially limiting the use of reporter molecules like GFP to estimate efficacy in early development stages. Moreover, the expression of reporter molecules may not reflect internalization efficiency, which can be pertinent information for the development of these systems. Therefore, as their popularity continues to rise, more sensitive and accurate tools will be necessary.
We validated a DNA extraction and qPCR assay to quantify the internalized plasmid copy number in NIH/3T3 cells transfected using LGA-PEI (lactic-co-glycolic acid-modified polyethylenimine) nanoparticles. Optimized qPCR and DNA extraction assays exhibited high linearity, sensitivity, and robustness. Additionally, we developed an appropriate extraction and extracellular plasmid digestion protocol that eliminated potential polymer interference with the qPCR reaction and increased specificity for internalized plasmid quantification.
Using these optimized protocols, we estimated that transfected NIH/3T3 cells incorporated approximately 9 500 to 150 000 plasmids/cell for transfection reactions that yielded as few as 4.1 %–18.2 % GFP-positive cells. Overall, these results indicate that our optimized method is reliable for quantifying transfected plasmids in context of low-efficiency nanoparticle carrier system.
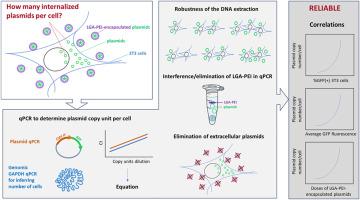
基于qpcr的非病毒纳米颗粒载体在贴壁细胞模型中质粒内化效率测定方法。
非病毒纳米颗粒介导的转染系统已在现代制药和生物技术应用中得到广泛接受,包括用于COVID-19疫苗的RNA递送。然而,这些系统可能表现出较低的转染和转录效率,潜在地限制了报告分子(如GFP)在早期发育阶段评估疗效的使用。此外,报告分子的表达可能不能反映内化效率,这可以为这些系统的开发提供相关信息。因此,随着它们的普及程度不断提高,将需要更灵敏、更准确的工具。我们验证了DNA提取和qPCR检测,以量化使用LGA-PEI(乳酸-羟基乙酸修饰的聚乙烯亚胺)纳米颗粒转染的NIH/3T3细胞的内化质粒拷贝数。优化后的qPCR和DNA提取分析具有较高的线性、灵敏度和鲁棒性。此外,我们开发了一种合适的提取和细胞外质粒消化方案,消除了qPCR反应中潜在的聚合物干扰,提高了内化质粒定量的特异性。使用这些优化的方案,我们估计转染的NIH/3T3细胞含有大约9500至150000个质粒/细胞,转染反应产生的gfp阳性细胞仅为4.1%至18.2%。总之,这些结果表明,我们优化的方法是可靠的定量转染质粒在低效率的纳米颗粒载体系统。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


