Structure of a functional archaellum in Bacteria of the Chloroflexota phylum
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
Motility in Archaea is driven by the archaellum, a rotary ATP-driven machinery unrelated to the bacterial flagellum. To date, archaella have been described exclusively in archaea; however, recent work reported archaellum genes in bacterial strains of the SAR202 clade (Chloroflexota). Here, using MacSyFinder, we show that bona fide archaellum gene clusters are widespread in several members of the Chloroflexota. Analysis of archaellum-encoding loci and Alphafold3-predicted structures show similarity to the archaellum machinery. Using cryo electron microscopy single-particle analysis, we solved the structure of the bacterial archaellum from Litorilinea aerophila to 2.7 Å. We also show the expression and assembly of this machinery in bacteria and its function in swimming motility. Finally, a phylogenomic analysis revealed two horizontal gene transfer events from euryarchaeal members to Chloroflexota. In summary, our study shows that a functional and assembled archaellum machinery can be exchanged between the two prokaryotic domains. Bona fide gene clusters for archaella are widespread across a bacterial phylum and encode functional motility machinery.
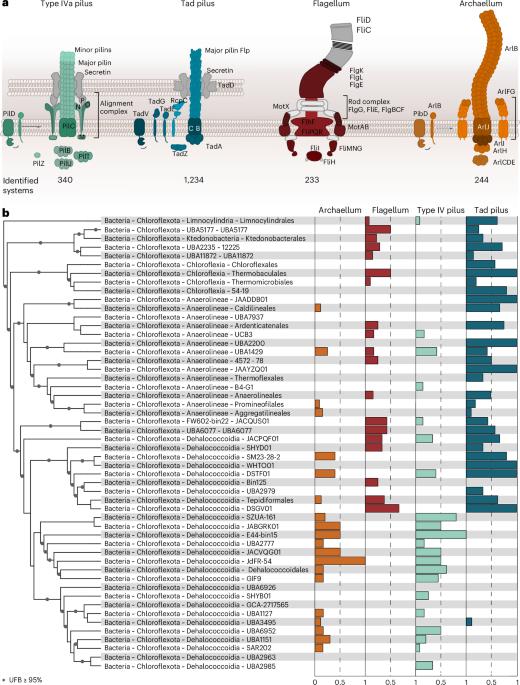
绿藻门细菌中一个功能性古菌的结构。
古细菌的运动是由古细菌驱动的,这是一种与细菌鞭毛无关的旋转atp驱动的机器。迄今为止,古菌只在古菌中被描述;然而,最近的工作报道了SAR202分支(氯氟菌)菌株中的古细菌基因。在这里,使用MacSyFinder,我们显示真正的古菌基因簇广泛存在于Chloroflexota的几个成员中。对古细菌编码位点的分析和alphafold3预测的结构显示出与古细菌的相似之处。利用低温电子显微镜单粒子分析,我们解出了Litorilinea aerophila至2.7 Å细菌古菌的结构。我们还展示了这种机制在细菌中的表达和组装及其在游泳运动中的作用。最后,系统基因组分析揭示了从euryarchaeal成员到Chloroflexota的两个水平基因转移事件。总之,我们的研究表明,一个功能性和组装的古细菌机器可以在两个原核结构域之间交换。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


