A highly sensitive magnetic separation-assisted electrochemical sensor for detection of m6A-microRNA-17–5p based on rolling circle amplification and N3-kethoxal labeling strategy at LIG electrodes
IF 6
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
N6-methyladenosine (m6A) is a prevalent epigenetic modification in eukaryotic RNA and exerts critical regulatory effects on the progression of cancers and other human disorders. Recent studies have revealed a close association between methylation levels in microRNA (miRNA) and human cancers, emphasizing the diagnostic potential of m6A-modified miRNAs for gastrointestinal (GI) cancers. However, the low abundance of m6A in miRNAs has hindered further research, highlighting the need for highly sensitive and selective detection methods capable of analyzing m6A in complex biological samples.
Results
We developed an innovative biosensor based on a dual amplification strategy—combining rolling circle amplification (RCA) with N3-kethoxal nucleic acid labeling—to enable ultrasensitive detection of m6A-microRNA-17–5p (m6A-RNA) at laser-induced graphene (LIG) electrodes. In the proposed strategy, m6A-RNA is quickly captured from the sample matrix using immunomagnetic beads and binds to the RCA primer–template complex. RCA is initiated under mild isothermal conditions, yielding long guanine-rich single-stranded DNAs (ssDNAs) that can be labeled with N3-kethoxal. Subsequently, a copper-free click reaction between N3-kethoxal and DBCO-biotin generates numerous binding sites for streptavidin-labeled horseradish peroxidase (SA-HRP), thereby catalyzing the electrochemical reaction system. The developed sensor demonstrated excellent sensitivity in the linear range of 0.1 pM–10 nM with a low detection limit of 10.1 fM, and performed well in selectivity, stability and reproducibility.
Significance
This study established a novel electrochemical signal amplification strategy, advancing the development of m6A modification analysis. Furthermore, the magnetic separation-assisted LIG electrode platform enables efficient target isolation from complex matrices while promoting device miniaturization. Successful detection of total RNA from MIA PaCa-2 cells and colorectal cancer (CRC) tissues demonstrates the method's applicability in clinical diagnostics, offering new avenues for GI cancer research and point-of-care biomarker testing.
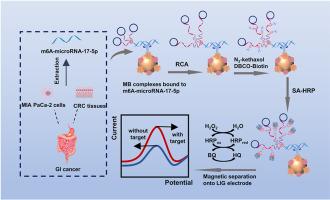
基于滚动圆放大和n3 -酮醛标记策略的高灵敏度磁分离辅助电化学传感器检测m6A-microRNA-17-5p
n6 -甲基腺苷(m6A)是真核生物RNA中普遍存在的表观遗传修饰,对癌症和其他人类疾病的进展具有重要的调控作用。最近的研究揭示了microRNA (miRNA)甲基化水平与人类癌症之间的密切联系,强调了m6a修饰的miRNA对胃肠道(GI)癌症的诊断潜力。然而,mirna中m6A的低丰度阻碍了进一步的研究,这突出了对能够分析复杂生物样品中m6A的高灵敏度和选择性检测方法的需求。我们开发了一种基于双扩增策略的创新生物传感器,将滚动圈扩增(RCA)与n3 -酮醛核酸标记相结合,可以在激光诱导石墨烯(LIG)电极上超灵敏地检测m6A-microRNA-17-5p (m6A-RNA)。在提出的策略中,使用免疫磁珠从样品基质中快速捕获m6A-RNA并结合到RCA引物-模板复合物上。RCA在温和的等温条件下启动,产生可以用n3酮醛标记的富含鸟嘌呤的长单链dna (ssdna)。随后,n3 -酮醛和dbco -生物素之间的无铜点击反应产生了许多链霉亲和素标记的辣根过氧化物酶(SA-HRP)的结合位点,从而催化了电化学反应体系。该传感器在0.1 pM ~ 10 nM的线性范围内具有良好的灵敏度,检测限低至10.1 fM,具有良好的选择性、稳定性和重复性。本研究建立了一种新的电化学信号放大策略,推动了m6A修饰分析的发展。此外,磁分离辅助LIG电极平台可以在促进器件小型化的同时,从复杂的矩阵中有效地分离目标。MIA PaCa-2细胞和结直肠癌(CRC)组织中总RNA的成功检测证明了该方法在临床诊断中的适用性,为GI癌症研究和即时生物标志物检测提供了新的途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


