Scaling up spatial transcriptomics for large-sized tissues: uncovering cellular-level tissue architecture beyond conventional platforms with iSCALE
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Recent advances in spatial transcriptomics (ST) technologies have transformed our ability to profile gene expression while preserving crucial spatial context within tissues. However, existing ST platforms are constrained by high costs, long turnaround times, low resolution, limited gene coverage and inherently small tissue capture areas, which hinder their broad applications. Here we present iSCALE, a method that reconstructs large-scale, super-resolution gene expression landscapes and automatically annotates cellular-level tissue architecture in samples exceeding capture areas of current ST platforms. The performance of iSCALE was assessed by comprehensive evaluations involving benchmarking experiments, immunohistochemistry staining and manual annotations by pathologists. When applied to multiple sclerosis human brain samples, iSCALE uncovered lesion-associated cellular characteristics undetectable by conventional ST experiments. Our results demonstrate the utility of iSCALE in analyzing large tissues by enabling unbiased annotation, resolving cell type composition, mapping cellular microenvironments and revealing spatial features beyond the reach of standard ST analysis or routine histopathological assessment. iSCALE leverages histology and spatial transcriptomics to infer gene expression at super resolution in large tissues.
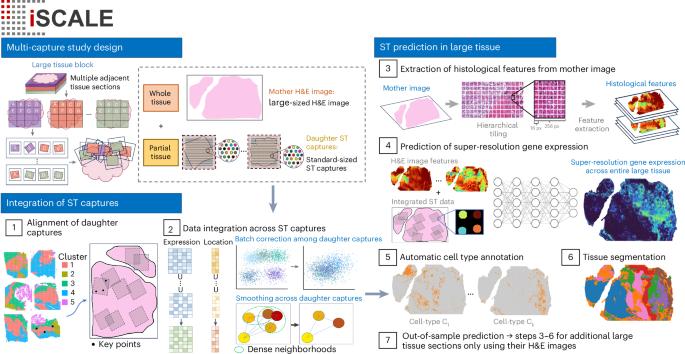
扩大大尺寸组织的空间转录组学:用iSCALE揭示超出传统平台的细胞水平组织结构。
空间转录组学(ST)技术的最新进展改变了我们在保留组织内关键空间背景的同时分析基因表达的能力。然而,现有的ST平台受到高成本、长周转时间、低分辨率、有限的基因覆盖和固有的小组织捕获区域的限制,阻碍了它们的广泛应用。在这里,我们提出了iSCALE,一种重建大规模,超分辨率基因表达景观的方法,并在超过当前ST平台捕获区域的样本中自动注释细胞水平的组织结构。通过基准实验、免疫组织化学染色和病理学家手工注释等综合评价来评估iSCALE的性能。当应用于多发性硬化症人脑样本时,iSCALE揭示了常规ST实验无法检测到的病变相关细胞特征。我们的研究结果证明了iSCALE在分析大组织方面的实用性,它可以实现无偏注释、解析细胞类型组成、绘制细胞微环境和揭示超出标准ST分析或常规组织病理学评估范围的空间特征。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


