Cancer subclone detection based on DNA copy number in single-cell and spatial omic sequencing data
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Somatic mutations such as copy number alterations accumulate during cancer progression, driving intratumor heterogeneity that impacts therapy effectiveness. Understanding the characteristics and spatial distribution of genetically distinct subclones is essential for unraveling tumor evolution and improving cancer treatment. Here we present Clonalscope, a subclone detection method using copy number profiles, applicable to spatial transcriptomics and single-cell sequencing data. Clonalscope implements a nested Chinese Restaurant Process to identify de novo tumor subclones, which can incorporate prior information from matched bulk DNA sequencing data for improved subclone detection and malignant cell labeling. On single-cell RNA sequencing and single-cell assay for transposase-accessible chromatin using sequencing data from gastrointestinal tumors, Clonalscope successfully labeled malignant cells and identified genetically different subclones with thorough validations. On spatial transcriptomics data from various primary and metastasized tumors, Clonalscope labeled malignant spots, traced subclones and identified spatially segregated subclones with distinct differentiation levels and expression of genes associated with drug resistance and survival. Clonalscope is a method for cancer subclone detection leveraging copy number profiles estimated using spatial and single-cell sequencing data.
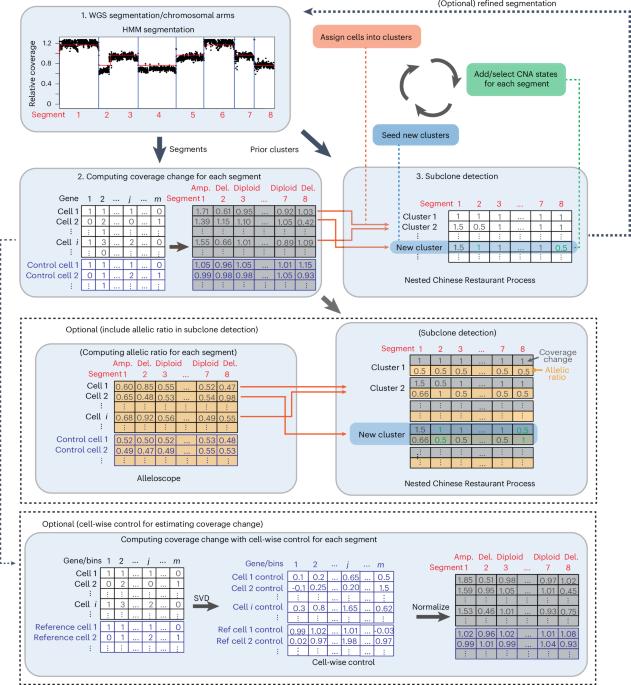
基于单细胞DNA拷贝数和空间组学测序数据的癌症亚克隆检测。
体细胞突变如拷贝数改变在癌症进展过程中积累,驱动影响治疗效果的肿瘤内异质性。了解遗传上不同亚克隆的特征和空间分布对于揭示肿瘤进化和改善癌症治疗至关重要。在这里,我们提出了Clonalscope,一种使用拷贝数谱的亚克隆检测方法,适用于空间转录组学和单细胞测序数据。Clonalscope实现了一个嵌套的中国餐馆过程来识别新生肿瘤亚克隆,该过程可以结合匹配的大量DNA测序数据的先验信息,以改进亚克隆检测和恶性细胞标记。在单细胞RNA测序和利用胃肠道肿瘤的测序数据进行转座酶可及染色质的单细胞检测中,Clonalscope成功地标记了恶性细胞,并通过彻底的验证鉴定了遗传上不同的亚克隆。根据来自各种原发和转移性肿瘤的空间转录组学数据,Clonalscope标记了恶性斑点,追踪了亚克隆,并鉴定了空间分离的亚克隆,这些亚克隆具有不同的分化水平和与耐药和生存相关的基因表达。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


