Controllable targeted protein degradation as a promising tool for discovery of novel cellular and developmental mechanisms
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
Abstract
Studying the spatial and temporal roles of essential proteins remains technically challenging. The effectiveness of perturbing gene functions using well established approaches upstream of the protein level, such as conditional knockout and RNA interference or morpholino-mediated knockdown, are often dependent upon the turnover rate of pre-existing proteins. Acute targeted protein degradation technologies can circumvent this limitation, and has emerged as powerful tools for discoveries of previously unrecognized protein functions in highly dynamic cellular and developmental processes. Auxin-inducible degron, degrade green fluorescent protein, degradation tag and proteolysis-targeting chimera are efficient for rapid knockdown of degron-tagged or untagged endogenous proteins in a controllable manner. All these approaches harness the evolutionarily conserved multi-protein E3 ubiquitin ligase complex in targeting proteins for degradation by the proteasome, offering versatile applications for protein functional studies in yeasts, plants, invertebrates, and vertebrates. This review presents the understanding of spatial and temporal protein functions advanced by commonly used auxin-inducible degron, degrade green fluorescent protein and degradation tag technologies. It also mentions the promising therapeutic potentials offered by the proteolysis-targeting chimera. With constant improvements, targeted protein degradation will open up new avenues for unprecedented findings of the dynamic features in a living system.
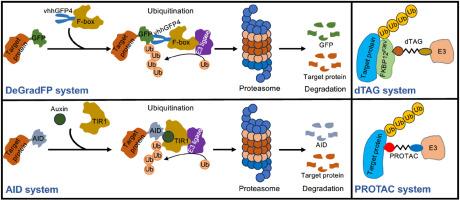
可控的靶向蛋白降解是发现新的细胞和发育机制的有前途的工具。
研究必需蛋白质的时空作用在技术上仍然具有挑战性。利用蛋白质水平上游的成熟方法(如条件敲除和RNA干扰或morpholino介导的敲低)干扰基因功能的有效性通常取决于先前存在的蛋白质的周转率。急性靶向蛋白质降解技术可以绕过这一限制,并已成为发现高度动态细胞和发育过程中以前未被识别的蛋白质功能的有力工具。生长素诱导的降解蛋白、降解绿色荧光蛋白、降解标签和蛋白水解靶向嵌合体能够以可控的方式快速敲除有降解标记或未标记的内源蛋白。所有这些方法都利用进化上保守的多蛋白E3泛素连接酶复合物来靶向蛋白质被蛋白酶体降解,为酵母、植物、无脊椎动物和脊椎动物的蛋白质功能研究提供了广泛的应用。本文综述了常用的生长素诱导降解、降解绿色荧光蛋白和降解标签技术对蛋白质时空功能的研究进展。它还提到了蛋白水解靶向嵌合体提供的有希望的治疗潜力。随着不断的改进,靶向蛋白质降解将为生命系统中前所未有的动态特征发现开辟新的途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


