Mapping transposon insertion sites within bacterial genomes by direct Sanger sequencing
IF 2.5
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
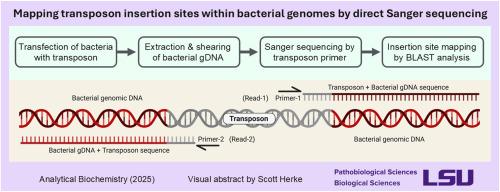
Microbial biomedical research frequently involves mutagenesis by insertion of transposons into the genome. Currently, transposon insertion locations are typically elucidated by Next Generation Sequencing or by Sanger sequencing of PCR products. Here, transposons were located by direct Sanger sequencing of bacterial genomic DNA from both Salmonella enterica (Gram-negative) and Staphylococcus aureus (Gram-positive) cultures. DNA was prepared by shearing to a modal size of ∼2 kb followed by purification by paramagnetic beads. Sequencing reactions involved relatively minor modifications of standard protocols (e.g., extra sequencing polymerase, 75–100 PCR cycles); completed reactions were cleaned by ethanol-EDTA precipitation. Reads were generated on the ABI 3130xl Genetic Analyzer using 50-cm capillary arrays and a run protocol modified for extra sample injection time. Good quality reads of ∼500–800 nt were routinely generated; BLAST results returned nearly 100% matches to genomes in the NCBI database. As implemented, the optimized protocol (post-DNA extraction) could be performed within an 8-h workday (with sequencing results the following day) for ∼$10 (USD) per sequencing reaction. Although this method was developed to locate transposons inserted into bacterial genomes, it seems likely that it can be extended to generate sequence data from even native single-copy genes from small genomes (e.g., <5 Mb).
通过直接Sanger测序在细菌基因组中定位转座子插入位点。
微生物生物医学研究经常涉及将转座子插入基因组的诱变。目前,转座子插入位置通常是通过下一代测序或PCR产物的Sanger测序来确定的。在这里,转座子是通过直接桑格测序从肠炎沙门氏菌(革兰氏阴性)和金黄色葡萄球菌(革兰氏阳性)培养的细菌基因组DNA定位。通过剪切至约2kb的模态大小制备DNA,然后用顺磁珠纯化。测序反应涉及对标准方案相对较小的修改(例如,额外的测序聚合酶,75-100个PCR循环);完成的反应用乙醇- edta沉淀法清洗。在ABI 3130xl遗传分析仪上使用50厘米毛细管阵列和修改的运行方案以增加样品注射时间。常规生成约500-800 nt的高质量读数;BLAST结果与NCBI数据库中的基因组几乎100%匹配。优化后的方案(dna后提取)可以在8小时工作日内完成(第二天有测序结果),每个测序反应约10美元。虽然这种方法是为了定位插入细菌基因组的转座子而开发的,但它似乎可以扩展到从小基因组(例如,
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


