Advancements in sample preparation and extraction techniques for metabolomic analysis of animal fluids
IF 4.9
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
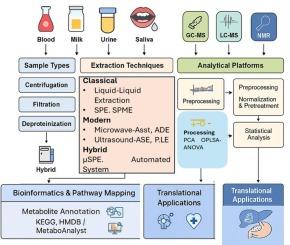
Metabolomics has become an extremely valuable tool in investigating biochemical processes in animal disease diagnosis, nutrition, and health. The precision of metabolomic data, however, is heavily dependent on sample preparation and extraction, which are still the most error-susceptible and variable steps in the workflow. Animal biofluids such as milk, urine, plasma, and serum pose especially challenging problems due to their complex protein, salt, and lipid matrices that make it difficult to detect metabolites. This review gives a systematic and critical evaluation of traditional, modern, and mixed extraction techniques, including protein precipitation, liquid–liquid extraction, solid-phase extraction, and microextraction-based methods such as SPME, DLLME, and QuEChERS. Mini case studies demonstrate their actual use in animal metabolomics, and provide context-dependent advantages, limitations, and method selection strategies. We also address the expanding role of hybrid and automated systems, including robotic platforms and microfluidics, to enhance reproducibility and scalability for large-cohort research. Analytical platform integration with GC–MS, LC–MS, CE–MS, and NMR is addressed, centring on platform selection criteria and complementary multi-platform approaches. Besides extraction, we address major areas of quality control, standardization, and inter-laboratory reproducibility, and then move to advances in data pre-processing, normalization, and pathway analysis based on bioinformatics. Finally, we touch on translational applications in animal and human disease, and outline future directions that integrate automation, artificial intelligence, and green extraction chemistries. Through the integration of methodological detail with pragmatic case examples, this review at once fulfills state-of-the-art summarization and forward-looking vision and offers pragmatic guidance for optimizing metabolomic investigations of animal fluids.
动物体液代谢组学分析的样品制备和提取技术进展
代谢组学已经成为研究动物疾病诊断、营养和健康中的生化过程的极有价值的工具。然而,代谢组学数据的准确性在很大程度上依赖于样品制备和提取,这仍然是工作流程中最容易出错和可变的步骤。动物生物液体,如牛奶、尿液、血浆和血清,由于其复杂的蛋白质、盐和脂质基质,使得检测代谢物变得困难,因此构成了特别具有挑战性的问题。本文综述了传统、现代和混合提取技术,包括蛋白质沉淀、液-液萃取、固相萃取和基于微萃取的方法,如SPME、DLLME和QuEChERS。小型案例研究展示了它们在动物代谢组学中的实际应用,并提供了上下文相关的优势、局限性和方法选择策略。我们还讨论了混合和自动化系统的扩展作用,包括机器人平台和微流体,以提高大型队列研究的可重复性和可扩展性。分析平台集成与GC-MS, LC-MS, CE-MS和NMR解决,集中在平台选择标准和互补的多平台方法。除了提取之外,我们还讨论了质量控制、标准化和实验室间可重复性的主要领域,然后转向数据预处理、规范化和基于生物信息学的途径分析方面的进展。最后,我们谈到了动物和人类疾病的转化应用,并概述了集成自动化,人工智能和绿色提取化学的未来方向。通过将方法细节与实际案例相结合,本综述立即实现了最先进的总结和前瞻性的愿景,并为优化动物体液代谢组学研究提供了实用的指导。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Microchemical Journal
化学-分析化学
CiteScore
8.70
自引率
8.30%
发文量
1131
审稿时长
1.9 months
期刊介绍:
The Microchemical Journal is a peer reviewed journal devoted to all aspects and phases of analytical chemistry and chemical analysis. The Microchemical Journal publishes articles which are at the forefront of modern analytical chemistry and cover innovations in the techniques to the finest possible limits. This includes fundamental aspects, instrumentation, new developments, innovative and novel methods and applications including environmental and clinical field.
Traditional classical analytical methods such as spectrophotometry and titrimetry as well as established instrumentation methods such as flame and graphite furnace atomic absorption spectrometry, gas chromatography, and modified glassy or carbon electrode electrochemical methods will be considered, provided they show significant improvements and novelty compared to the established methods.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


