DiameTR: A cryo-EM tool for diameter sorting of tubular samples
IF 5.1
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
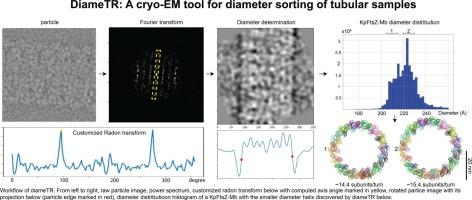
Tubular structures are ubiquitous in biological systems and have been a focal point of cryo-electron microscopy (cryo-EM) structural analysis since the technique’s inception. A critical step in processing tubular cryo-EM data is particle classification by diameter, as uniformity in diameter is a prerequisite for high-resolution three-dimensional reconstructions. Conventional methods rely on cross-correlation-based algorithms, which require prior knowledge to generate reference images, or iterative two-dimensional (2D) classification, that align and cluster particles into a predefined number of classes—a process that is both time-consuming and subjective. To address these limitations, we developed diameTR, a computational tool that rapidly determines tubular diameters in a prior knowledge-free and reference-free manner using GPU-accelerated processing on a per-particle basis. When applied to homogeneous datasets, diameTR yields narrow diameter distributions aligning closely with published values. For heterogeneous samples, it enables the separation of subsets with distinct diameters, validated by 2D averaging. Notably, diameTR identified an unreported smaller diameter subset of particles with new helical symmetry parameters in the previously published KpFtsZ-Monobody dataset. Collectively, diameTR represents a robust, efficient solution for diameter determination in tubular cryo-EM samples, eliminating the need for extensive human intervention while significantly accelerating processing.
DiameTR:用于管状样品直径分选的低温电镜工具
管状结构在生物系统中无处不在,自低温电子显微镜(cryo-EM)技术问世以来,一直是低温电子显微镜(cryo-EM)结构分析的焦点。处理管状低温电镜数据的关键步骤是按直径对颗粒进行分类,因为直径的均匀性是高分辨率三维重建的先决条件。传统的方法依赖于基于交叉相关的算法,这需要先验知识来生成参考图像,或者依赖于迭代二维(2D)分类,将粒子排列并聚集到预定义的类别中,这是一个既耗时又主观的过程。为了解决这些限制,我们开发了diameTR,这是一种计算工具,可以使用gpu加速处理,以无先验知识和无参考的方式快速确定管材直径。当应用于同质数据集时,diameter r产生的窄直径分布与公布的值非常接近。对于异质样本,它可以分离具有不同直径的子集,通过二维平均验证。值得注意的是,在之前发表的KpFtsZ-Monobody数据集中,diameTR发现了一个未报道的具有新螺旋对称参数的直径较小的粒子子集。总的来说,diameTR代表了一种强大、高效的解决方案,用于管状冷冻电镜样品的直径测定,消除了大量人为干预的需要,同时显著加快了处理速度。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Structural Biology: X
Biochemistry, Genetics and Molecular Biology-Structural Biology
CiteScore
6.50
自引率
0.00%
发文量
20
审稿时长
62 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


