Structural analysis of ASCH domain-containing proteins and their implications for nucleotide processing
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
ASC-1 homology (ASCH) domain family proteins are believed to play essential roles in RNA metabolism, but detailed structural and functional information is limited. Research has shown that the E. coli enzyme YqfB, which contains an ASCH domain, has amidohydrolase activity, converting N4-acetylcytidine (ac4C) RNA nucleoside into cytidine. Here, we present the crystal structures of EcYqfB both in its unbound state and bound to a substrate. Our analysis reveals how the substrate interacts with the enzyme, offering insights into its catalytic mechanism. In vivo experiments further show that deleting EcYqfB does not change overall ac4C levels across various RNA types, indicating that EcYqfB specifically functions in ac4C nucleoside metabolism. We also determined the structures of two homologous proteins: mouse EOLA1 and the human TRIP4-ASCH domain, highlighting differences in their substrate preferences. These findings offer important insights for future research into the structure and function of the ASCH domain protein family.
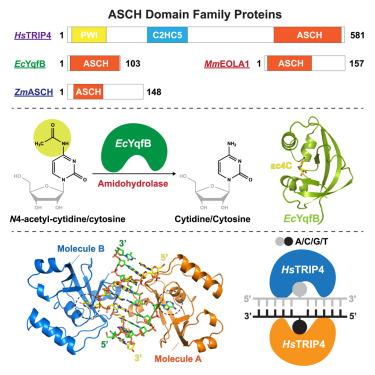
含ASCH结构域蛋白的结构分析及其对核苷酸加工的意义
ASC-1同源性(ASCH)结构域家族蛋白被认为在RNA代谢中起重要作用,但详细的结构和功能信息有限。研究表明,含有ASCH结构域的大肠杆菌酶YqfB具有氨基水解酶活性,可将n4 -乙酰胞苷(ac4C) RNA核苷转化为胞苷。在这里,我们展示了EcYqfB在其非结合状态和与底物结合状态下的晶体结构。我们的分析揭示了底物如何与酶相互作用,为其催化机制提供了见解。体内实验进一步表明,删除EcYqfB不会改变各种RNA类型中ac4C的总体水平,这表明EcYqfB特异性地在ac4C核苷代谢中起作用。我们还确定了两个同源蛋白的结构:小鼠EOLA1和人类TRIP4-ASCH结构域,突出了它们对底物偏好的差异。这些发现为进一步研究ASCH结构域蛋白家族的结构和功能提供了重要的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


