Initial leads to combat streptogramin resistance generated from X-ray fragment screening against VatD
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Streptogramins are potent antibiotics targeting bacterial ribosome. The synergistic binding of group A and B streptogramins to 50S-ribosome yields bactericidal effects. However, their efficacy is compromised by resistance mechanisms, including enzymatic acetylation of group A streptogramins by virginiamycin acetyltransferase (Vat) enzymes, which reduces their affinity for ribosomes. Using fragment-based drug discovery we identified starting points for development of VatD inhibitors. X-ray crystallography screening revealed three primary fragment-binding sites on VatD. In the acetyl-binding subsite, fragments stabilized distinct conformational states in critical residues, His82 and Trp121. In the antibiotic-binding site, two fragments formed interactions that could be leveraged for competitive inhibition. Elaborations of these fragments showed weak inhibition of VatD activity, indicating potential for further optimization. These findings establish initial hits that could restore streptogramin efficacy by targeting VatD directly, providing a structural foundation for inhibitor development against resistant bacterial strains.
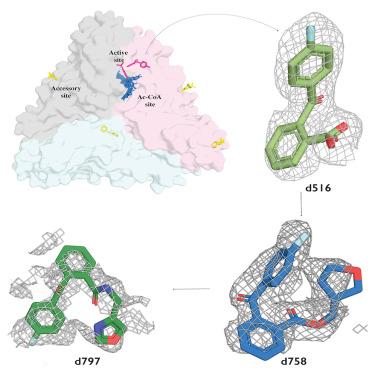
针对VatD的x射线片段筛选产生的链状gramin耐药性的初步结果
链状gramins是针对细菌核糖体的有效抗生素。A组和B组链状蛋白与50s核糖体的协同结合产生杀菌作用。然而,它们的功效受到抗性机制的影响,包括弗吉尼亚霉素乙酰转移酶(Vat)酶对A族链霉素的酶促乙酰化,从而降低了它们对核糖体的亲和力。利用基于片段的药物发现,我们确定了VatD抑制剂开发的起点。x射线晶体学筛选显示VatD上有三个主要的片段结合位点。在乙酰基结合亚位点,片段稳定了关键残基His82和Trp121的不同构象状态。在抗生素结合位点,两个片段形成相互作用,可以用于竞争性抑制。这些片段的修饰显示出对VatD活性的弱抑制,表明进一步优化的潜力。这些发现建立了可以通过直接靶向VatD来恢复链状gramin疗效的初始点,为开发针对耐药菌株的抑制剂提供了结构基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


