Expression and function of new candidate regulators of placodal neurogenesis in Xenopus laevis
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
Abstract
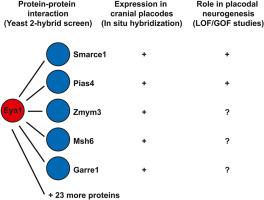
The transcription factor Six1 and its co-activator Eya1 play central and varied roles during the development of sensory neurons derived from the cranial placodes in vertebrates. Previous studies suggested that these proteins promote both the maintenance of proliferative neuronal progenitors and neuronal differentiation. Context-specific interactions of Six1 and/or Eya1 with different cofactors are likely to contribute to the activation of distinct target genes during different stages of placodal neurogenesis. However, while some protein binding partners of Six1 have recently been identified, little is known about Eya1 interaction partners. To uncover additional Eya1 interaction partners with potential roles in placodal neurogenesis, we here conduct a yeast two-hybrid screen in Xenopus laevis. Apart from confirming Six1 (Six1.L) and Six4 (Six4.L and Six4.S) as known protein interaction partners of Eya1, this screen identified 25 additional candidate binding partners including several transcription factors, chromatin modifiers and enzymes involved in posttranslational modifications. Analysis of the expression of genes encoding five of these candidates (garre1, msh6, zmym3, pias4, smarce1) during X. laevis embryogenesis by in situ hybridization, revealed extensive co-expression of each of these with eya1 during placodal development. This suggests potential roles of these candidates in the regulation of placodal neurogenesis possibly in conjunction with Eya1. In gain and loss of function studies of the E3 SUMO-ligase Pias4 and the chromatin modifier Smarce1, we confirm essential roles of both of these proteins for placodal neurogenesis but their mode of interaction with Eya1 remains to be further clarified.
非洲爪蟾胎盘神经发生新候选调控因子的表达和功能。
转录因子Six1及其共激活因子Eya1在脊椎动物源自颅基板的感觉神经元的发育过程中发挥着核心和多种作用。先前的研究表明,这些蛋白既促进维持增殖的神经元祖细胞,又促进神经元分化。Six1和/或Eya1与不同辅助因子的上下文特异性相互作用可能有助于在胎盘神经发生的不同阶段激活不同的靶基因。然而,尽管最近已经发现了Six1的一些蛋白质结合伙伴,但对Eya1的相互作用伙伴知之甚少。为了发现在胎盘神经发生中具有潜在作用的其他Eya1相互作用伙伴,我们在非洲爪蟾中进行了酵母双杂交筛选。除了确认Six1和Six4是Eya1的两个已知蛋白相互作用伙伴外,该筛选还确定了25个候选结合伙伴,包括几种转录因子,染色质修饰剂和参与翻译后修饰的酶。通过原位杂交分析了5个候选基因(garre1, msh6, zmym3, pias4, smarce1)在青松胚胎发生过程中的表达,发现它们在胎盘发育过程中与eya1广泛共表达。这表明这些候选基因在调节胎盘神经发生中的潜在作用可能与Eya1共同作用。在E3 sumo -连接酶Pias4和染色质修饰剂Smarce1的功能获得和丧失研究中,我们证实了这两种蛋白在胎盘神经发生中的重要作用,但它们与Eya1的相互作用模式仍有待进一步阐明。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


