A kinetic and spectroscopic study of tetrahydrodipicolinate N-succinyltransferase (DapD) from Serratia marcescens and its inactivation by Cu2+
IF 3
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
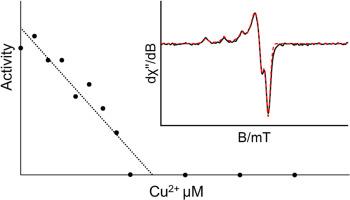
Tetrahydrodipicolinate N-succinyltransferase (DapD) catalyzes the reaction of tetrahydrodipicolinate (THDP) and succinyl-CoA to form (S)-2-(3-carboxypropanamido)-6-oxoheptanedioic acid and coenzyme A. The enzyme is in the diaminopimelate-lysine biosynthesis pathway which produces two metabolites necessary for the survival and growth of pathogenic bacteria. Since lysine is an essential amino acid to humans, DapD is a potentially safe target for antibiotic therapies. Despite its identification as an exploitable target, details regarding the mechanism of DapD and its inhibition remain poorly resolved. In this work, the DAPD gene from Serratia marcescens has been optimally expressed in E. coli host cells and purified. Initial velocity patterns generated using two assays (one direct and one coupled) are consistent with a rapid equilibrium ordered bi bi kinetic mechanism. Therefore, conversion of the central enzyme complexes (chemistry) is thought to be the rate-limiting step in the reaction. Data fitted to the appropriate rate equation provide estimates of kinetic constants for the reaction (Kia (2-AP) = 1.9 ± 0.26 mM and Kb (succinyl-CoA) = 87 ± 15 μM). SmDapD is rapidly and specifically inactivated in the presence of Cu2+ at low concentrations, yet activity was protected in a reducing environment (1 mM DTT). Correspondingly, the internal tryptophan fluorescence of DapD is quenched by Cu2+ with a KD of 2.7 μM, and emission spectra are comparable to chemically denatured DapD. Fluorescence quenching was relieved in the presence of DTT or EDTA. EPR spectra are consistent with Cu2+ binding to enzymatic histidine and being reduced to Cu+. These data suggest Cu2+ binding to the enzyme as well as an oxidative inactivation that results in significant structural changes and denaturing. Together, the mechanistic detail and characterization of the Cu2+ inactivation will inform the targeting of DapD with new antibiotic therapies.
粘质沙雷菌四氢二吡啶酸n -琥珀基转移酶(DapD)的动力学和光谱研究及其Cu2灭活。
四氢二吡啶酸n -琥珀酰转移酶(DapD)催化四氢二吡啶酸(THDP)与琥珀酰辅酶a反应生成(S)-2-(3-羧基丙胺)-6-氧七烷二酸和辅酶a。该酶在二氨基苯甲酸-赖氨酸生物合成途径中产生致病菌生存和生长所必需的两种代谢物。由于赖氨酸是人类必需的氨基酸,因此DapD是抗生素治疗的潜在安全靶点。尽管它被认为是一个可利用的靶点,但关于DapD的机制及其抑制的细节仍然很不清楚。在这项工作中,粘质沙雷氏菌的DAPD基因在大肠杆菌宿主细胞中得到了最佳表达和纯化。用两种测定法(一种直接测定法和一种耦合测定法)生成的初速度模式与快速平衡有序bi - bi动力学机制一致。因此,中心酶复合物的转化(化学)被认为是反应中的限速步骤。拟合相应速率方程的数据给出了反应的动力学常数(Kia (2-AP) = 1.9±0.26 mM, Kb (succinyl-CoA) = 87±15 μM)。SmDapD在低浓度Cu2+存在下迅速特异性失活,但在还原环境(1mm DTT)下活性得到保护。相应的,DapD内部色氨酸荧光被Cu2+猝灭,KD为2.7 μM,发射光谱与化学变性的DapD相当。在DTT或EDTA的存在下,荧光猝灭得以缓解。EPR光谱显示Cu2+与酶促组氨酸结合并被还原为Cu+。这些数据表明Cu2+与酶结合以及氧化失活导致显著的结构变化和变性。总之,Cu2+失活的机制细节和特征将为新的抗生素治疗靶向DapD提供信息。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Archives of biochemistry and biophysics
生物-生化与分子生物学
CiteScore
7.40
自引率
0.00%
发文量
245
审稿时长
26 days
期刊介绍:
Archives of Biochemistry and Biophysics publishes quality original articles and reviews in the developing areas of biochemistry and biophysics.
Research Areas Include:
• Enzyme and protein structure, function, regulation. Folding, turnover, and post-translational processing
• Biological oxidations, free radical reactions, redox signaling, oxygenases, P450 reactions
• Signal transduction, receptors, membrane transport, intracellular signals. Cellular and integrated metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


