Scvi-hub: an actionable repository for model-driven single-cell analysis
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
The growing availability of single-cell omics datasets presents new opportunities for reuse, while challenges in data transfer, normalization and integration remain a barrier. Here we present scvi-hub: a platform for efficiently sharing and accessing single-cell omics datasets using pretrained probabilistic models. It enables immediate execution of fundamental tasks like visualization, imputation, annotation and deconvolution on new query datasets using state-of-the-art methods, with massively reduced storage and compute requirements. We show that pretrained models support efficient analysis of large references, including the CZI CELLxGENE Discover Census. Scvi-hub is built within the scvi-tools open-source environment and integrated into scverse. Scvi-hub offers a scalable and user-friendly framework for accessing and contributing to a growing ecosystem of ready-to-use models and datasets, thus putting the power of atlas-level analysis at the fingertips of a broad community of users. Scvi-hub is a versatile and efficient platform for model-based analysis of single-cell sequencing studies with access to a diverse array of datasets and downstream analysis.
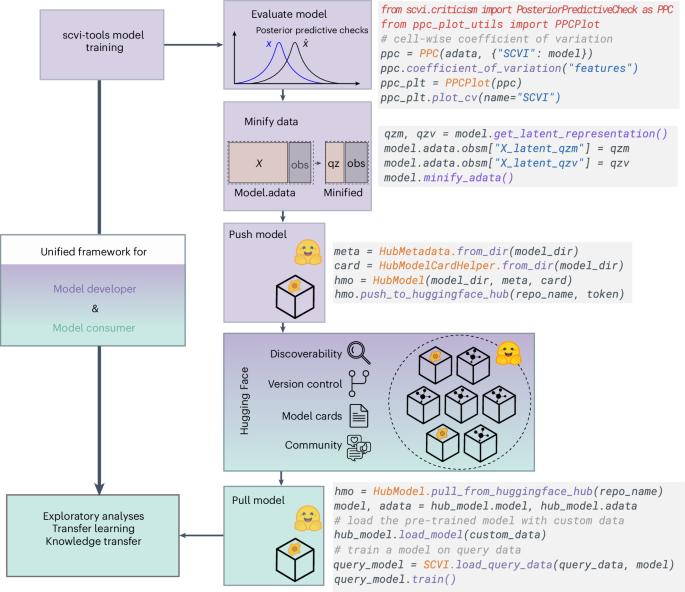
Scvi-hub:用于模型驱动的单细胞分析的可操作存储库。
单细胞组学数据集的日益可用性为重用提供了新的机会,而数据传输、规范化和集成方面的挑战仍然是一个障碍。在这里,我们提出scvi-hub:一个使用预训练概率模型有效共享和访问单细胞组学数据集的平台。它可以使用最先进的方法在新的查询数据集上立即执行基本任务,如可视化、imputation、注释和反卷积,同时大大减少了存储和计算需求。我们表明,预训练模型支持对大型参考文献的有效分析,包括CZI CELLxGENE Discover Census。Scvi-hub构建在scvi-tools开源环境中,并集成到scverse中。Scvi-hub提供了一个可扩展和用户友好的框架,用于访问和贡献不断增长的即用型模型和数据集生态系统,从而将atlas级分析的功能置于广泛的用户社区的指尖。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


