Culture-independent meta-pangenomics enabled by long-read metagenomics reveals associations with pediatric undernutrition
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The human gut microbiome is linked to child malnutrition, yet traditional microbiome approaches lack resolution. We hypothesized that complete metagenome-assembled genomes (cMAGs), recovered through long-read (LR) DNA sequencing, would enable pangenome and microbial genome-wide association study (GWAS) analyses to identify microbial genetic associations with child linear growth. LR methods produced 44–64× more cMAGs per gigabase pair (Gbp) than short-read methods, with PacBio (PB) yielding the most accurate and cost-effective assemblies. In a Malawian longitudinal pediatric cohort, we generated 986 cMAGs (839 circular) from 47 samples and applied this database to an expanded set of 210 samples. Machine learning identified species predictive of linear growth. Pangenome analyses revealed microbial genetic associations with linear growth, while genome instability correlated with declining length-for-age Z score (LAZ). This resource demonstrates the power of comparing cMAGs with health trajectories and establishes a new standard for microbiome association studies.
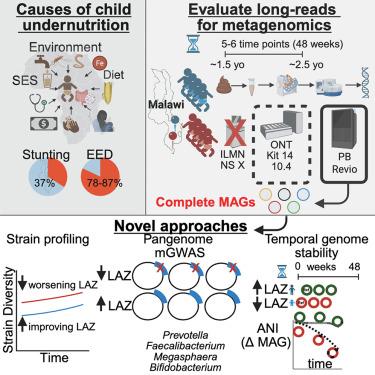
由长读元基因组学实现的培养独立元泛基因组学揭示了与儿科营养不良的关联
人类肠道微生物组与儿童营养不良有关,但传统的微生物组方法缺乏解决方案。我们假设,通过长读(LR) DNA测序恢复的完整宏基因组组装基因组(cMAGs)将使泛基因组和微生物全基因组关联研究(GWAS)分析能够确定微生物遗传与儿童线性生长的关联。LR方法比短读方法每千兆酶对(Gbp)多产生44 - 64倍的cMAGs, PacBio (PB)产生最准确和最经济的组装。在马拉维纵向儿科队列中,我们从47个样本中生成了986个cmagg(839个圆形),并将该数据库应用于扩展的210个样本。机器学习识别物种预测线性增长。泛基因组分析显示,微生物遗传与线性生长相关,而基因组不稳定性与年龄长度Z评分(LAZ)下降相关。该资源证明了将cmag与健康轨迹进行比较的能力,并为微生物组关联研究建立了新的标准。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


