HPDAF: A practical tool for predicting drug-target binding affinity using multimodal features
IF 5.9
2区 医学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
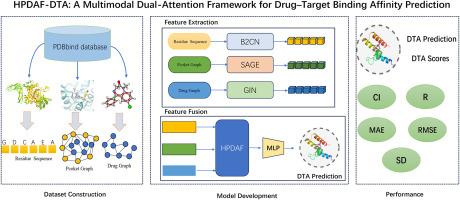
Accurate prediction of drug-target binding affinity is crucial for efficient drug discovery and design, enabling researchers to better understand molecular interactions and accelerate the identification of promising drug candidates. Despite recent advances, existing computational methods often face difficulties in effectively combining detailed structural information from drug-binding sites with broader molecular features. Here, we introduce HPDAF, a practical multimodal deep learning tool designed to improve the accuracy of drug-target binding affinity predictions. HPDAF uniquely integrates three types of biochemical information: protein sequences, drug molecular graphs, and structural interaction data from protein-binding pockets. Each of these data types is carefully processed using specialized modules that capture essential molecular characteristics. A novel hierarchical attention-based mechanism then effectively combines these diverse features, enabling the model to dynamically emphasize the most relevant structural and sequential information. Extensive evaluations using widely recognized benchmark datasets, including CASF-2016 and CASF-2013, demonstrate that HPDAF consistently achieves superior predictive performance compared to current state-of-the-art methods. The practical applicability and enhanced accuracy of HPDAF highlight its potential as a valuable computational tool for medicinal chemists involved in drug design and virtual screening efforts.
HPDAF:使用多模态特征预测药物-靶标结合亲和力的实用工具
准确预测药物靶标结合亲和力对于有效的药物发现和设计至关重要,使研究人员能够更好地了解分子相互作用并加速有希望的候选药物的鉴定。尽管最近取得了进展,但现有的计算方法在有效地将药物结合位点的详细结构信息与更广泛的分子特征结合起来时往往面临困难。在这里,我们介绍HPDAF,一个实用的多模态深度学习工具,旨在提高药物靶点结合亲和力预测的准确性。HPDAF独特地集成了三种类型的生化信息:蛋白质序列,药物分子图和蛋白质结合口袋的结构相互作用数据。每种数据类型都使用捕获基本分子特征的专门模块进行仔细处理。然后,一种新的基于注意力的分层机制有效地结合了这些不同的特征,使模型能够动态地强调最相关的结构和顺序信息。使用广泛认可的基准数据集(包括CASF-2016和CASF-2013)进行的广泛评估表明,与当前最先进的方法相比,HPDAF始终具有卓越的预测性能。HPDAF的实用性和增强的准确性突出了它作为药物化学家参与药物设计和虚拟筛选工作的有价值的计算工具的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
11.70
自引率
9.00%
发文量
863
审稿时长
29 days
期刊介绍:
The European Journal of Medicinal Chemistry is a global journal that publishes studies on all aspects of medicinal chemistry. It provides a medium for publication of original papers and also welcomes critical review papers.
A typical paper would report on the organic synthesis, characterization and pharmacological evaluation of compounds. Other topics of interest are drug design, QSAR, molecular modeling, drug-receptor interactions, molecular aspects of drug metabolism, prodrug synthesis and drug targeting. The journal expects manuscripts to present the rational for a study, provide insight into the design of compounds or understanding of mechanism, or clarify the targets.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


