Multigenerational proteolytic inactivation of restriction upon subtle genomic hypomethylation in Pseudomonas aeruginosa
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
Restriction-modification (R-M) systems protect against phage infection by detecting and degrading invading foreign DNA. However, like many prokaryotic anti-phage defences, R-M systems pose a major risk of autoimmunity, exacerbated by the presence of hundreds to thousands of potential cleavage sites in the bacterial genome. Pseudomonas aeruginosa strains experience the temporary inactivation of restriction endonucleases following growth at high temperatures, but the reason and mechanisms for this phenomenon are unknown. Here we report that P. aeruginosa type I restriction endonuclease is degraded and the methyltransferase is partially degraded, by two Lon-like proteases when replicating at >41 °C. This post-translational regulation prevents self-DNA targeting, which is a risk due to stable genomic hypomethylation, as demonstrated by single-molecule, real-time sequencing and TadA-assisted N6-methyladenosine sequencing. When cells grown at >41 °C are returned to 37 °C, full genomic methylation does not fully recover for up to 60 bacterial generations, and thus restriction activity remains off for the duration. Our findings demonstrate that type I R-M is tightly regulated post-translationally with a long memory effect that ensures genomic stability and mitigates autotoxicity. Elevated temperatures inactivate a type I restriction endonuclease in P. aeruginosa for ~60 generations via proteolytic degradation triggered by DNA hypomodification, preventing self-targeting and enabling recovery of restriction activity.
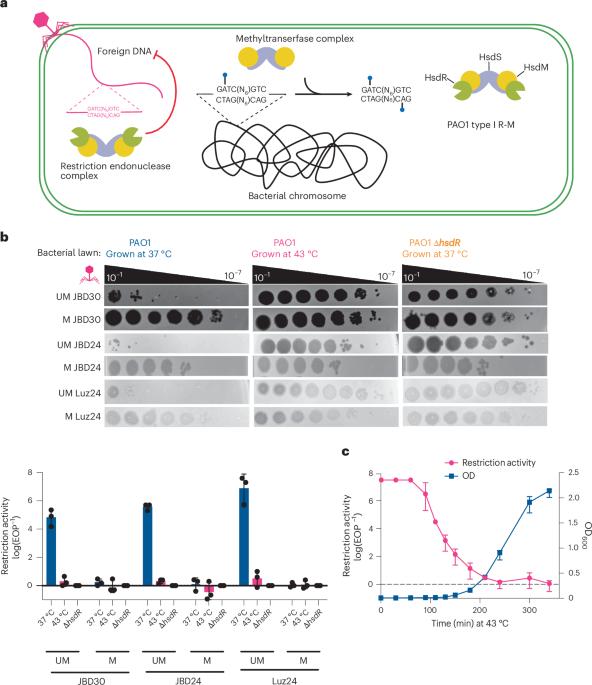
铜绿假单胞菌基因组低甲基化限制的多代蛋白水解失活
限制性修饰(R-M)系统通过检测和降解入侵的外源DNA来防止噬菌体感染。然而,像许多原核抗噬菌体防御一样,R-M系统具有自身免疫的主要风险,并且由于细菌基因组中存在数百到数千个潜在的切割位点而加剧。铜绿假单胞菌菌株在高温下生长后会经历限制性内切酶的暂时失活,但这种现象的原因和机制尚不清楚。在这里,我们报道了P. aeruginosa I型限制性内切酶被降解,甲基转移酶被两个lon样蛋白酶部分降解,当在41°C下复制时。单分子实时测序和tada辅助的n6 -甲基腺苷测序证明,这种翻译后调控阻止了自dna靶向,这是由于稳定的基因组低甲基化而造成的风险。当在41°C下生长的细胞返回到37°C时,完整的基因组甲基化在60代细菌中不会完全恢复,因此限制性酶活性在此期间保持关闭。我们的研究结果表明,I型R-M在翻译后受到严格调控,具有长记忆效应,确保基因组稳定性并减轻自毒性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


