Unraveling the developmental heterogeneity of human retinal ganglion cells within the developing retina to study to the continuity of maturation
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
Abstract
Tissue development is a complex spatiotemporal process with multiple interdependent components. Anatomical, histological, sequencing, and evolutional strategies can be used to profile and explain tissue development from different perspectives. The introduction of single-cell RNA sequencing (scRNAseq) methods and the computational tools allows to deconvolute developmental heterogeneity and draw a decomposed uniform map. In this manuscript, we decomposed the development of a human retina with a focus on the retinal ganglion cells (RGC). To increase the temporal resolution of retinal cell classes maturation state we assumed the working hypothesis that that maturation of retinal ganglion cells is a continuous, non-discrete process. We have assembled the scRNAseq atlas of human fetal retina from fetal week 8 to week 27 and applied the computational methods to unravel maturation heterogeneity into a uniform maturation track. We align RGC transcriptomes in pseudotime to map RGC developmental fate trajectories against the broader timeline of retinal development. Through this analysis, we identified the continuous maturation track of RGC and described the cell-intrinsic (differentially expressed and variable genes, maturation gene profiles, regulons, transcriptional motifs) and -extrinsic profiles (neurotrophic receptors across maturation, cell-cell interactions) of different RGC maturation states. We described the genes involved in the retina and RGC maturation, including de novo RGC maturation drivers. We demonstrate the application of the human fetal retina atlas as a reference tool, allowing automated annotation and universal embedding of scRNAseq data. Altogether, our findings deepen the current knowledge of the retina and RGC maturation by bringing in the maturation dimension for the cell class vs. state analysis. We show how the pseudotime application contributes to developmental-oriented analyses, allowing to order the cells by their maturation state. This approach not only improves the downstream computational analysis but also provides a true maturation track transcriptomics profile.
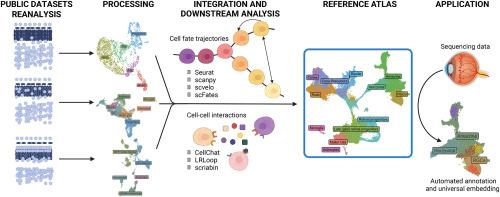
揭示发育中的视网膜神经节细胞的发育异质性,以研究成熟的连续性。
组织发育是一个复杂的时空过程,具有多个相互依赖的组成部分。解剖、组织学、测序和进化策略可用于从不同角度描述和解释组织发育。单细胞RNA测序(scRNAseq)方法和计算工具的引入允许反卷积发育异质性并绘制分解的均匀图谱。在这篇文章中,我们分解了人类视网膜的发育,重点是视网膜神经节细胞(RGC)。为了提高视网膜细胞类别成熟状态的时间分辨率,我们假设视网膜神经节细胞的成熟是一个连续的、非离散的过程。我们收集了从胎儿第8周到第27周的人胎儿视网膜的scRNAseq图谱,并应用计算方法将成熟异质性分解为统一的成熟轨迹。我们将RGC转录组在伪时间上对齐,以绘制RGC发育命运轨迹与视网膜发育的更广泛时间轴。通过这一分析,我们确定了RGC的连续成熟轨迹,并描述了不同RGC成熟状态的细胞内在特征(差异表达和可变基因、成熟基因谱、调控、转录基序)和外在特征(跨成熟的神经营养受体、细胞-细胞相互作用)。我们描述了参与视网膜和RGC成熟的基因,包括新生的RGC成熟驱动因素。我们演示了人类胎儿视网膜图谱作为参考工具的应用,允许scRNAseq数据的自动注释和通用嵌入。总之,我们的发现通过引入成熟维度用于细胞类别与状态分析,加深了目前对视网膜和RGC成熟的了解。我们展示了伪时间应用程序如何有助于面向发育的分析,允许按细胞的成熟状态排序。这种方法不仅改善了下游的计算分析,而且提供了一个真正的成熟轨迹转录组学图谱。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


