Mismatch-sensitive DNA hybridization controlled by inchworm-type peptide nucleic acid–PEG conjugates
IF 2.5
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
The duplex-forming behavior of an inchworm-type PNA–PEG conjugate (i-PPc), engineered for the selective recognition of point mutations in DNA, was assessed through thermodynamic analysis employing UV melting curves and circular dichroism spectroscopy. The i-PPc demonstrated the ability to form stable duplexes exclusively with fully complementary DNA sequences, while no hybridization with single-base mismatched sequences. This binary on/off hybridization behavior was maintained even under physiologically relevant conditions (37 °C), thereby illustrating the exceptional point mutation discrimination capability of i-PPc. The behavior observed can be ascribed to the distinctive structure of i-PPc, wherein two PNA segments, possessing intrinsically different duplex-forming stabilities—high and low—are covalently linked via a flexible PEG linker. The high-stability PNA segment functions as the primary recognition domain for point mutations, thereby defining the sequence specificity of duplex formation. Conversely, the low-stability segment contributes cooperatively to the overall duplex stabilization only when the high-stability segment successfully hybridizes with the target DNA. This cooperative mechanism underlies the sequence-selective duplex formation of i-PPc, highlighting its potential as a highly specific probe for DNA mutation diagnostics. These findings indicate that i-PPc represents a promising platform for point mutation detection and nucleic acid-based molecular diagnostics grounded in DNA hybridization under physiological conditions.
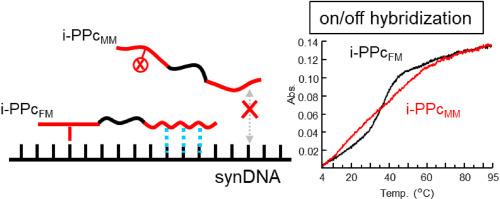
尺蠖型肽核酸- peg偶联物控制错配敏感DNA杂交
采用紫外熔化曲线和圆二色光谱热力学分析评估了一种用于选择性识别DNA点突变的尺蠖型PNA-PEG共轭物(i-PPc)的双工形成行为。i-PPc能够与完全互补的DNA序列形成稳定的双链,而与单碱基不匹配的序列不杂交。即使在生理相关条件下(37°C),这种二元开/关杂交行为也能保持,从而说明i-PPc具有特殊的点突变识别能力。观察到的行为可以归因于i-PPc的独特结构,其中两个具有本质上不同的双工形成稳定性的PNA片段-高和低-通过柔性PEG连接共价连接。高稳定性的PNA片段作为点突变的主要识别域,从而定义了双链形成的序列特异性。相反,只有当高稳定性片段成功地与目标DNA杂交时,低稳定性片段才有助于整体双工稳定。这种合作机制是i-PPc序列选择性双工形成的基础,突出了其作为DNA突变诊断的高度特异性探针的潜力。这些发现表明,在生理条件下,i-PPc为点突变检测和基于DNA杂交的核酸分子诊断提供了一个很有前景的平台。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


