Exploration of endoplasmic reticulum stress-related gene markers in amyotrophic lateral sclerosis: a comprehensive analysis of bioinformatics and machine learning
IF 2.5
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
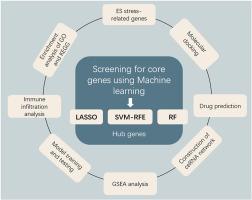
This study aimed to investigate potential biomarkers related to Endoplasmic reticulum (ER) stress in Amyotrophic lateral sclerosis (ALS) through a comprehensive bioinformatic approach. The gene expression profiles of ALS patients and healthy controls were downloaded from the Gene Expression Omnibus (GEO) database. ER stress-related genes were collected from the MSigDB databases and document literature. The “limma” R package was employed to detect the differentially expressed ER stress-related genes (DE-ERSGs). Three methods of machine learning were applied to select the hub DE-ERSGs. ROC curves were conducted to evaluate model performance. An external dataset was chosen to evaluate the diagnostic capability of hub genes. The CIBERSORT algorithm was used to evaluate the immune cell infiltration characteristics. Additionally, we constructed a systematic ceRNA regulatory network using Cytoscape software and predicted the possible drug candidates using the Enrichr platform. Molecular docking analysis was used to further validate the binding ability of the candidate drug molecules to the hub genes. Six hub DE-ERSGs (ABCA1, CKAP4, TOR1AIP1, MMP9, EDC4, and ALPP) were identified, and the related models performed well. These hub genes were concentrated in multiple pathways and related to various immune cells. Drugs such as nitroglycerin, diazepam, FENRETINIDE, and edaravone exhibited good binding affinity to the hub genes, indicating that they may be promising drugs for the management of ALS. This study revealed the essential role of ER stress in the pathogenesis of ALS from an integrative perspective, providing guidance for the development of new therapeutic targets and diagnostic strategies.
肌萎缩侧索硬化症内质网应激相关基因标记的探索:生物信息学和机器学习的综合分析
本研究旨在通过综合生物信息学方法研究肌萎缩侧索硬化症(ALS)内质网(ER)应激相关的潜在生物标志物。从gene expression Omnibus (GEO)数据库下载ALS患者和健康对照者的基因表达谱。从MSigDB数据库和文献文献中收集内质网应激相关基因。“limma”R包检测内质网应激相关基因(de - ergs)的差异表达。采用三种机器学习方法选择轮毂de - ergs。采用ROC曲线评价模型的性能。选择一个外部数据集来评估枢纽基因的诊断能力。采用CIBERSORT算法评价免疫细胞浸润特性。此外,我们使用Cytoscape软件构建了一个系统的ceRNA调控网络,并使用enrichment平台预测了可能的候选药物。通过分子对接分析进一步验证候选药物分子与枢纽基因的结合能力。确定了6个hub de - ergs (ABCA1、CKAP4、TOR1AIP1、MMP9、EDC4和ALPP),相关模型表现良好。这些枢纽基因集中在多种途径中,与多种免疫细胞有关。硝酸甘油、地西泮、芬拉啶和依达拉奉等药物与中枢基因表现出良好的结合亲和力,表明它们可能是治疗ALS的有希望的药物。本研究从综合角度揭示了内质网应激在ALS发病机制中的重要作用,为开发新的治疗靶点和诊断策略提供指导。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


