Single-cell analysis of genetically minimized Salmonella reveals effector gene cooperation in vivo
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
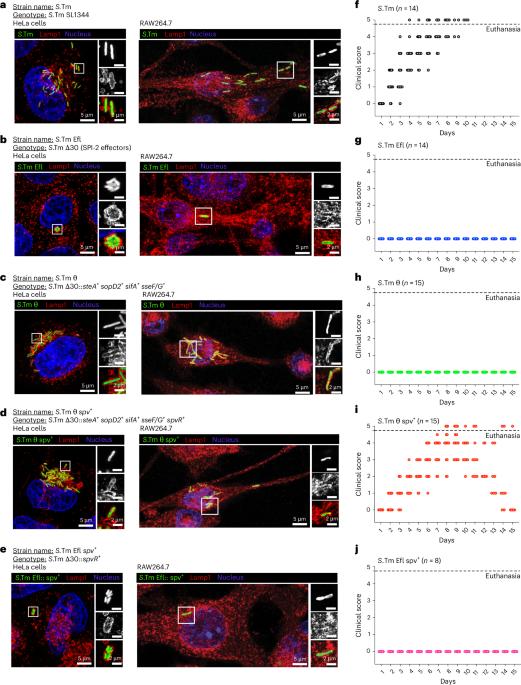
Bacterial pathogens such as Salmonella Typhimurium (S.Tm) can deliver large repertoires of effector proteins directly into host cells. Due to the genetic and functional redundancies found in these systems, it has been difficult to determine how individual effector proteins cooperate with one another to elicit pathogenic phenotypes in vivo. To overcome this challenge, targeted genome minimization has been used to identify small effector protein networks sufficient for S.Tm colonization of complex tissues. Here we used mass cytometry to generate a comprehensive map of minimal effector network functions at single-cell resolution. This approach revealed the temporal progression of S.Tm transmission and uncovered a CD62L+ monocyte population as a major bottleneck to cell-to-cell dissemination in the spleen. We further show how cooperation between two effector gene networks acquired during distinct episodes of bacterial evolution has shaped the cellular and tissue tropism of non-typhoidal Salmonella species. A minimal effector subset enables Salmonella Typhimurium to overcome bottlenecks regulated by the early innate immune response and establish infection within a CD62L+ monocyte niche in the spleen.
单细胞分析基因最小化沙门氏菌揭示效应基因在体内的合作
像鼠伤寒沙门氏菌这样的细菌病原体可以将大量的效应蛋白直接传递到宿主细胞中。由于在这些系统中发现的遗传和功能冗余,很难确定个体效应蛋白如何相互合作以在体内引发致病表型。为了克服这一挑战,靶向基因组最小化已被用于鉴定足以使S.Tm定植于复杂组织的小效应蛋白网络。在这里,我们使用大量细胞术在单细胞分辨率下生成最小效应网络功能的综合图。该方法揭示了S.Tm传播的时间进展,并发现CD62L+单核细胞群是脾脏细胞间传播的主要瓶颈。我们进一步展示了在细菌进化的不同时期获得的两个效应基因网络之间的合作如何塑造了非伤寒沙门氏菌物种的细胞和组织趋向性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


