A base editor facilitates simultaneous purine and pyrimidine substitutions for ex vivo and in vivo mutagenesis screens
IF 7.2
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Genetic mutations are closely linked to human diseases, yet the relationship between many mutations and their corresponding phenotypes remains poorly understood. Furthermore, tools to study the connection between nucleotide variations and phenotypes are limited. To address this issue, we developed ACGBEmax by fusing the dual-functional deaminase, engineered N-methylpurine DNA glycosylase, and evolved SOS response associated peptidase domain with nCas9(D10A). ACGBEmax enables the precise conversion of A, C, and G to other bases in mammalian cells, thereby generating an extensive range of base mutations types. We used ACGBEmax to generate HPRT variants, identifying mutations conferring resistance to 6-thioguanine. Additionally, we performed in situ mutagenesis of Ctnnb1 in mouse liver, identifying both known and potential oncogenic mutations. Our results prove that ACGBEmax is a powerful tool for generating a wide spectrum of mutation types at specific gene loci, highlighting its significant potential for applications in functional screening and the directed evolution of protein variants.
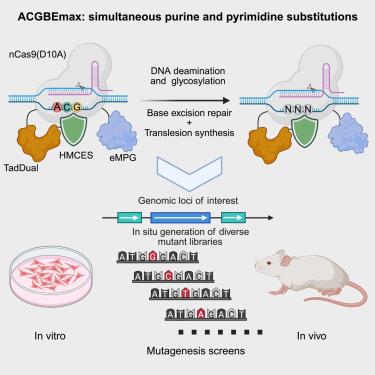
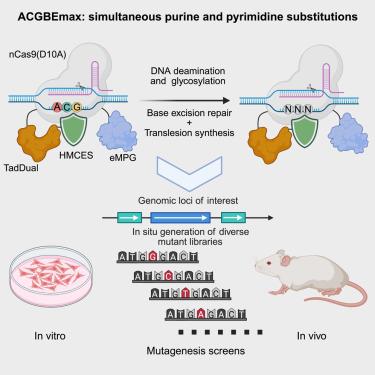
碱基编辑器促进体外和体内突变筛选同时进行嘌呤和嘧啶替换
基因突变与人类疾病密切相关,但许多突变与其相应表型之间的关系仍然知之甚少。此外,研究核苷酸变异和表型之间联系的工具是有限的。为了解决这一问题,我们通过融合双功能脱氨酶,工程化n -甲基嘌呤DNA糖基化酶,并进化出与nCas9(D10A)相关的SOS反应相关肽酶结构域,开发出ACGBEmax。ACGBEmax能够将A、C和G精确地转化为哺乳动物细胞中的其他碱基,从而产生广泛的碱基突变类型。我们使用ACGBEmax生成HPRT变体,鉴定对6-硫鸟嘌呤产生抗性的突变。此外,我们在小鼠肝脏中对Ctnnb1进行了原位诱变,确定了已知和潜在的致癌突变。我们的研究结果证明,ACGBEmax是一个强大的工具,可以在特定基因位点上产生广泛的突变类型,突出了它在功能筛选和蛋白质变异的定向进化方面的巨大潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell Chemical Biology
Biochemistry, Genetics and Molecular Biology-Molecular Medicine
CiteScore
14.70
自引率
2.30%
发文量
143
期刊介绍:
Cell Chemical Biology, a Cell Press journal established in 1994 as Chemistry & Biology, focuses on publishing crucial advances in chemical biology research with broad appeal to our diverse community, spanning basic scientists to clinicians. Pioneering investigations at the chemistry-biology interface, the journal fosters collaboration between these disciplines. We encourage submissions providing significant conceptual advancements of broad interest across chemical, biological, clinical, and related fields. Particularly sought are articles utilizing chemical tools to perturb, visualize, and measure biological systems, offering unique insights into molecular mechanisms, disease biology, and therapeutics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


