Mutation profiling, evolution analysis, molecular dynamics simulation, and functional characterization of Omicron sub-strains
IF 2.7
4区 医学
Q3 VIROLOGY
引用次数: 0
Abstract
The ongoing mutation and evolution of SARS-CoV-2 have posed a severe threat to global health, and their functional impact remains to be further characterized. Here, we analyzed the selection pressure from 49 Omicron sub-strains at the gene and amino acid levels. We also examined the impact of mutations on the binding affinity between the receptor binding domain (RBD) and angiotensin-Converting Enzyme 2 (ACE2) and evaluated the immune escape ability of RBD responding to the monoclonal antibodies (mAbs) through molecular dynamics simulation on eight representative Omicron sub-variants (B.1.1.529, BA.2, XBB.1.5, BA.2.86, JN.1, KP.2, KP.3, and KP.3.1.1). We identified 12 positive selection mutation sites on the viral S protein, including 11 mutation sites in the N-terminal domain (NTD) and RBD regions. A large number of accumulated mutation sites led to an increase in the receptor binding affinity of B.1.1.529 and BA.2.86. In particular, the “saltatory” evolution of BA.2.86 reached to its maximum binding affinity. The E484K mutation exhibited the highest binding affinity in the BA.2.86 and its descendants. New mutation sites either did not affect the binding affinity (R346T, L455S and F456L) or decrease the affinity (K356T and Q493E), reflecting the fluctuation of total receptor binding force. Mutations and shortened conformational epitopes on RBD may mediate the immune escape in the variants of BA.2.86. Moreover, we revealed that the ABBV-47D11 monoclonal antibody could widely bind to the RBD mutation sites of various mutant strains. Our findings may help understand the evolution of SARS-CoV-2 variants and develop novel strategies against SARS-CoV-2 infection.
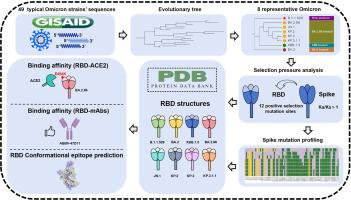
基因突变谱、进化分析、分子动力学模拟和功能表征。
SARS-CoV-2的持续突变和进化对全球健康构成了严重威胁,其功能影响有待进一步研究。在基因和氨基酸水平上分析了49个Omicron亚菌株的选择压力。我们还研究了突变对受体结合域(RBD)与血管紧张素转换酶2 (ACE2)结合亲和力的影响,并通过分子动力学模拟8个代表性的Omicron亚变体(B.1.1.529、BA.2、XBB.1.5、BA.2.86、j .1、KP.2、KP.3和KP.3.1.1)评估了RBD对单克隆抗体(mab)的免疫逃逸能力。我们在病毒S蛋白上发现了12个阳性选择突变位点,其中11个突变位点位于n端结构域(NTD)和RBD区域。大量突变位点的积累导致B.1.1.529和BA.2.86的受体结合亲和力增加。特别是BA.2.86的“跳跃式”进化达到了其最大的结合亲和力。E484K突变在BA.2.86及其后代中表现出最高的结合亲和力。新的突变位点要么不影响结合亲和力(R346T、L455S和F456L),要么降低亲和力(K356T和Q493E),反映了总受体结合力的波动。RBD上的突变和缩短的构象表位可能介导BA.2.86变异体的免疫逃逸。此外,我们发现ABBV-47D11单克隆抗体可以广泛结合各种突变株的RBD突变位点。我们的发现可能有助于了解SARS-CoV-2变异的演变,并制定对抗SARS-CoV-2感染的新策略。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Virus research
医学-病毒学
CiteScore
9.50
自引率
2.00%
发文量
239
审稿时长
43 days
期刊介绍:
Virus Research provides a means of fast publication for original papers on fundamental research in virology. Contributions on new developments concerning virus structure, replication, pathogenesis and evolution are encouraged. These include reports describing virus morphology, the function and antigenic analysis of virus structural components, virus genome structure and expression, analysis on virus replication processes, virus evolution in connection with antiviral interventions, effects of viruses on their host cells, particularly on the immune system, and the pathogenesis of virus infections, including oncogene activation and transduction.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


