Altered expression patterns of lncRNA MEG3 and LINC01611 in patients with colorectal cancer
IF 0.7
Q4 GENETICS & HEREDITY
引用次数: 0
Abstract
Colorectal cancer (CRC) is a major global health challenge, with long non-coding RNAs (lncRNAs) gaining attention as potential diagnostic biomarkers. This study aimed to experimentally validate bioinformatics findings on the expression patterns of maternally expressed gene 3 (MEG3) and LINC01611 in CRC patients from a specific ethnic population while considering associated risk factors. This in vitro study initially recruited 50 patients from a single ethnic group, with 48 completing the analysis after the exclusion of two samples. Two lncRNAs, MEG3 and LINC01611, were selected using Gene Expression Omnibus (GEO) microarray data and identified via R/BioConductor. Paired tissue samples (tumor and adjacent margins) were collected during surgery, and RNAs were extracted. Demographic and clinical data of patients were recorded, and gene expression was analyzed using quantitative real-time PCR (qPCR), with GAPDH as the internal control. Data analysis was performed using GraphPad Prism and SPSS software, with the significance level set at P < 0.05. The mean age of the patients was 59.5 ± 3.53 years, with 58.3 % (n = 28) being male, and 37.5 % of the patients had a history of smoking. The majority of patients had poorly differentiated (41.7 %) and stage II tumor (43.8 %), with lymph node metastasis commonly observed (60.4 %). The Wilcoxon signed-rank test revealed significant downregulation of MEG3 (32.396 fold change)and LINC01611(38.923 fold change) in tumor tissues compared to adjacent margins. A family history of CRC was associated with higher expression levels of MEG3 (1.48-fold, P = 0.038) and LINC01611 (1.03-fold, P = 0.007) in both tumor and margin tissues. Multivariable regression analysis demonstrated that lncRNAs had a significant association with tumor differentiation (P < 0.05), while other variables showed no statistically significant association (P > 0.05). Also, positive correlations were observed between MEG3 and LINC01611 expression levels in tumor (r = 0.649, P < 0.001) and margin (r = 0.424, P = 0.003) tissues. The significant downregulation of MEG3 and LINC01611 in tumor tissues compared to adjacent margin tissues highlights their potential role as tumor suppressors in CRC. These findings support further investigation into these lncRNAs as diagnostic biomarkers.
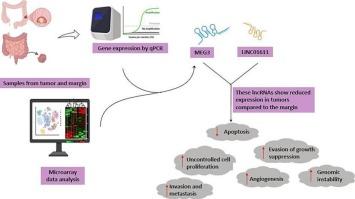
lncRNA MEG3和LINC01611在结直肠癌患者中的表达模式改变
结直肠癌(CRC)是一项重大的全球健康挑战,长链非编码rna (lncRNAs)作为潜在的诊断生物标志物越来越受到关注。本研究旨在通过实验验证母系表达基因3 (MEG3)和LINC01611在特定民族结直肠癌患者中的表达模式的生物信息学发现,同时考虑相关危险因素。这项体外研究最初从单一种族中招募了50名患者,其中48名在排除两个样本后完成了分析。通过GEO微阵列数据筛选MEG3和LINC01611两个lncrna,并通过R/BioConductor进行鉴定。术中收集成对组织样本(肿瘤及邻近边缘),提取rna。记录患者的人口学及临床资料,以GAPDH为内对照,采用实时荧光定量PCR (qPCR)分析基因表达情况。数据分析采用GraphPad Prism和SPSS软件,显著性水平设为P <; 0.05。患者平均年龄59.5±3.53岁,男性58.3% (n = 28), 37.5%有吸烟史。大多数患者为低分化(41.7%)和II期肿瘤(43.8%),常观察到淋巴结转移(60.4%)。Wilcoxon sign -rank检验显示,肿瘤组织中MEG3(32.396倍变化)和LINC01611(38.923倍变化)与邻近边缘相比显著下调。结直肠癌家族史与肿瘤和边缘组织中MEG3(1.48倍,P = 0.038)和LINC01611(1.03倍,P = 0.007)的高表达水平相关。多变量回归分析显示lncRNAs与肿瘤分化有显著相关性(P < 0.05),其他变量无统计学意义(P > 0.05)。MEG3与LINC01611在肿瘤组织(r = 0.649, P < 0.001)和切缘组织(r = 0.424, P = 0.003)中的表达水平呈正相关。与邻近边缘组织相比,MEG3和LINC01611在肿瘤组织中的显著下调突出了它们在结直肠癌中作为肿瘤抑制因子的潜在作用。这些发现支持进一步研究这些lncrna作为诊断性生物标志物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Human Gene
Biochemistry, Genetics and Molecular Biology (General), Genetics
CiteScore
1.60
自引率
0.00%
发文量
0
审稿时长
54 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


