A de novo-originated gene drives rose scent diversification
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
How evolution builds genes, how these genes attain enhanced expression, and how they integrate into existing regulatory networks to drive phenotypic diversification are all fascinating questions. Here, we generated chromosome-level genome assemblies for two Rosa banksiae subspecies and re-sequenced an additional 40 rose accessions. Genomic analysis of more than 100 Rosa accessions revealed multiple evolutionary steps leading to the de novo origination of a taxon-restricted gene, SCREP, specific to the rose lineage. Extensive transcriptomic, metabolomic, and functional analyses demonstrated that the recruitment of a Miniature Inverted-repeat Transposable Element (MITE) transposon into the gene promoter led to elevated expression, that the gene SCREP orchestrates eugenol biosynthesis, and that the evolutionary dynamics of SCREP account for variation in rose scent among different species and cultivars. Our results provide insights into the mechanism of de novo gene origination, the role of transposable elements in gene expression, and the evolutionary consequences of taxon-restricted genes in phenotypic diversification.
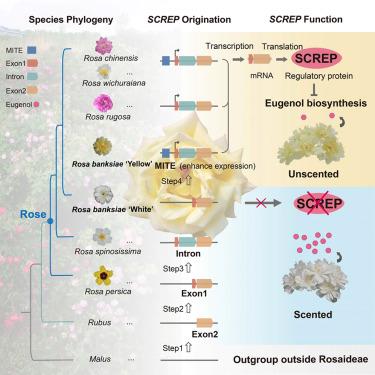
一种新产生的基因驱动了玫瑰香味的多样化
进化如何构建基因,这些基因如何获得增强表达,以及它们如何整合到现有的调控网络中以驱动表型多样化,这些都是令人着迷的问题。在这里,我们生成了两个banksiae亚种的染色体水平基因组组装,并对另外40个玫瑰材料进行了重新测序。对100多份玫瑰材料的基因组分析揭示了导致玫瑰谱系特有的分类群限制基因(SCREP)从头开始的多个进化步骤。广泛的转录组学、代谢组学和功能分析表明,微型反重复转座元件(micro - inver- repeat Transposable Element, MITE)转座子进入基因启动子导致表达升高,基因SCREP协调丁香酚的生物合成,并且SCREP的进化动力学解释了不同物种和栽培品种之间玫瑰香味的差异。我们的研究结果揭示了基因新生的机制,转座因子在基因表达中的作用,以及分类群限制基因在表型多样化中的进化后果。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


