Preventing CpG hypermethylation in oocytes safeguards mouse development
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
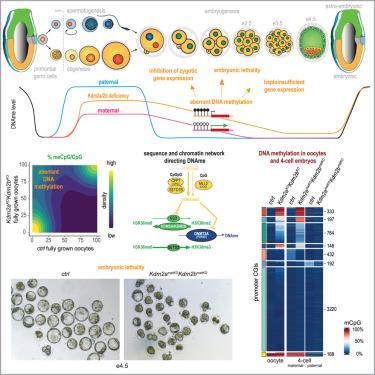
Except for regulatory CpG-island sequences, genomes of most mammalian cells are widely DNA-methylated. In oocytes, though, DNA methylation (DNAme) is largely confined to transcribed regions. The mechanisms restricting de novo DNAme in oocytes and their relevance thereof for zygotic genome activation and embryonic development are largely unknown. Here we show that KDM2A and KDM2B, two histone demethylases, prevent genome-wide accumulation of histone H3 lysine 36 di-methylation, thereby impeding DNMT3A-catalyzed DNAme. We demonstrate that aberrant DNAme at CpG islands inherited from Kdm2a/Kdm2b double-mutant oocytes represses gene transcription in two-cell embryos. Aberrant maternal DNAme impairs pre-implantation embryonic development, which is suppressed by Dnmt3a deficiency during oogenesis. Hence, KDM2A/KDM2B are essential for confining the oocyte methylome, thereby conferring competence for early embryonic development. Our research implies that the reprogramming capacity eminent to early embryos is insufficient for erasing aberrant DNAme from maternal chromatin, and that early development is susceptible to gene dosage haplo-insufficiency effects.
防止卵母细胞CpG超甲基化可保护小鼠发育
除了调节性cpg岛序列外,大多数哺乳动物细胞的基因组都被广泛的dna甲基化。然而,在卵母细胞中,DNA甲基化(DNAme)主要局限于转录区域。限制卵母细胞新生DNAme的机制及其与合子基因组激活和胚胎发育的相关性在很大程度上是未知的。本研究表明,两种组蛋白去甲基化酶KDM2A和KDM2B可阻止组蛋白H3赖氨酸36二甲基化的全基因组积累,从而阻碍dnmt3a催化的dna。我们证明,遗传自Kdm2a/Kdm2b双突变卵母细胞的CpG岛上的dna异常抑制了双细胞胚胎中的基因转录。异常的母体dna会损害着床前胚胎的发育,而胚胎发育在卵发生过程中受到Dnmt3a缺乏的抑制。因此,KDM2A/KDM2B对于限制卵母细胞甲基组至关重要,从而赋予早期胚胎发育的能力。我们的研究表明,早期胚胎的重编程能力不足以消除母体染色质中的异常dna,并且早期发育容易受到基因剂量单倍不足的影响。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


