Structural insights into outer membrane protein biogenesis in pathogenic Neisseria
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
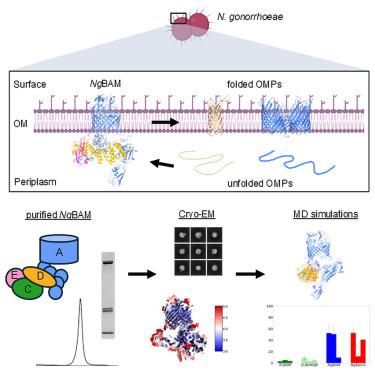
N. gonorrhoeae (Ngo) causes the sexually transmitted infection gonorrhea with ∼106 million infections worldwide annually. Ngo infections can result in an increased risk of acquiring HIV, infertility, and blindness. To combat Ngo infections, we report the cryoelectron microscopy (cryo-EM) structure of the Ngo β-barrel assembly machinery (NgBAM), which is responsible for the biogenesis of β-barrel outer membrane proteins (OMPs). NgBAM was observed in an inward-open state; however, the polypeptide transport-associated (POTRA) domains more closely match those found in the outward-open state in E. coli β-barrel assembly machinery (BAM). The barrel seam of NgBamA consists of partial pairing of strand β1 with β16; no outward-open state of NgBAM was observed. Molecular dynamics (MD) simulations reveal unique overall dynamics and interplay between the POTRA domains of NgBamA and NgBamD. We propose that in Ngo, initial recognition occurs in the inward-open state where the last strand of the OMP partially pairs with β1 of NgBamA and must compete off β16.
致病性奈瑟菌外膜蛋白生物发生的结构见解
淋病奈瑟菌(Ngo)引起性传播感染淋病,全世界每年感染约1.06亿例。非政府组织感染可能导致感染艾滋病毒、不孕症和失明的风险增加。为了对抗Ngo感染,我们报道了Ngo β-桶组装机制(NgBAM)的低温电镜(cryo-EM)结构,该机制负责β-桶外膜蛋白(OMPs)的生物发生。观察到NgBAM处于内向开放状态;然而,多肽转运相关结构域(POTRA)与大肠杆菌β-桶状组装机械(BAM)中外开放状态的结构域更接近。NgBamA的桶状缝由β1链与β16链的部分配对组成;未观察到NgBAM的外开状态。分子动力学(MD)模拟揭示了NgBamA和NgBamD的POTRA结构域之间独特的整体动力学和相互作用。我们认为,在Ngo中,最初的识别发生在内向开放状态,此时OMP的最后一条链与NgBamA的β1部分配对,并且必须与β16竞争。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


