Metabarcoding characterization of gastrointestinal strongyle nematodes in captive Asian elephants (Elephas maximus) and white rhinoceroses (Ceratotherium simum) in a private zoo, Thailand
IF 2.6
4区 医学
Q3 INFECTIOUS DISEASES
引用次数: 0
Abstract
Gastrointestinal strongyle nematodes pose significant health risks to captive megaherbivores, including Asian elephants (Elephas maximus) and white rhinoceroses (Ceratotherium simum). Traditional diagnostic methods often fail to accurately identify species due to morphological similarities, limiting understanding of parasite diversity and host-specificity. This study is among the first in Southeast Asia to apply high-throughput internal transcribed spacer-2 (ITS-2) rDNA metabarcoding to characterize strongyle nematode communities in these endangered hosts. Fecal samples from six rhinoceroses and four elephants housed in a private zoo in Thailand were processed using flotation, larval culture, and DNA extraction protocols. Amplicon sequencing was conducted on the Illumina MiSeq platform, and taxonomic assignments were performed using the DADA2 pipeline and NCBI/GenBank databases. Our results revealed the presence of strongyle infections. Murshidia spp. were detected in both host species, while Kiluluma ceratotherii was found exclusively in rhinoceroses. Phylogenetic analysis based on ITS-2 rDNA sequences demonstrated clear host-associated clades and suggested potential cryptic species within Kiluluma and Murshidia lineages. These findings provide new genetic evidence of host specificity and evolutionary divergence among strongylid nematodes in captive wildlife. The study underscores the utility of DNA metabarcoding for non-invasive parasite surveillance and highlights the urgent need to expand molecular databases for better taxonomic resolution in wildlife parasitology.
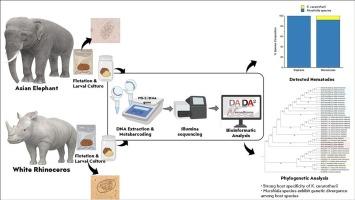
泰国一家私人动物园圈养亚洲象(大象)和白犀牛(白犀牛)胃肠道圆形线虫的元条形码特征
胃肠道圆形线虫对圈养的大型食草动物,包括亚洲象(象)和白犀牛(角犀)构成重大健康风险。由于形态相似性,传统的诊断方法往往不能准确识别物种,限制了对寄生虫多样性和宿主特异性的理解。该研究是东南亚首次应用高通量内转录间隔-2 (ITS-2) rDNA元条形码来表征这些濒危宿主的线虫群落。泰国一家私人动物园的6头犀牛和4头大象的粪便样本采用浮选、幼虫培养和DNA提取方案进行了处理。扩增子测序在Illumina MiSeq平台上进行,分类鉴定使用DADA2管道和NCBI/GenBank数据库。我们的结果显示存在圆形细胞感染。在两种宿主物种中均检测到Murshidia,而Kiluluma ceratotherii仅在犀牛中发现。基于ITS-2 rDNA序列的系统发育分析显示了与宿主相关的分支,并提出了Kiluluma和Murshidia谱系中潜在的隐种。这些发现为圈养野生动物中线虫的宿主特异性和进化分化提供了新的遗传证据。该研究强调了DNA元条形码在非侵入性寄生虫监测中的应用,并强调了扩大分子数据库以提高野生寄生虫学分类分辨率的迫切需要。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Infection Genetics and Evolution
医学-传染病学
CiteScore
8.40
自引率
0.00%
发文量
215
审稿时长
82 days
期刊介绍:
(aka Journal of Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases -- MEEGID)
Infectious diseases constitute one of the main challenges to medical science in the coming century. The impressive development of molecular megatechnologies and of bioinformatics have greatly increased our knowledge of the evolution, transmission and pathogenicity of infectious diseases. Research has shown that host susceptibility to many infectious diseases has a genetic basis. Furthermore, much is now known on the molecular epidemiology, evolution and virulence of pathogenic agents, as well as their resistance to drugs, vaccines, and antibiotics. Equally, research on the genetics of disease vectors has greatly improved our understanding of their systematics, has increased our capacity to identify target populations for control or intervention, and has provided detailed information on the mechanisms of insecticide resistance.
However, the genetics and evolutionary biology of hosts, pathogens and vectors have tended to develop as three separate fields of research. This artificial compartmentalisation is of concern due to our growing appreciation of the strong co-evolutionary interactions among hosts, pathogens and vectors.
Infection, Genetics and Evolution and its companion congress [MEEGID](http://www.meegidconference.com/) (for Molecular Epidemiology and Evolutionary Genetics of Infectious Diseases) are the main forum acting for the cross-fertilization between evolutionary science and biomedical research on infectious diseases.
Infection, Genetics and Evolution is the only journal that welcomes articles dealing with the genetics and evolutionary biology of hosts, pathogens and vectors, and coevolution processes among them in relation to infection and disease manifestation. All infectious models enter the scope of the journal, including pathogens of humans, animals and plants, either parasites, fungi, bacteria, viruses or prions. The journal welcomes articles dealing with genetics, population genetics, genomics, postgenomics, gene expression, evolutionary biology, population dynamics, mathematical modeling and bioinformatics. We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. Please click here for more information on our author services .
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


