Metagenomic and Molecular Simulation Insights into Plastic Degradation: Microenvironmental Matching and the Key Role of Residue Phe392
IF 11.3
1区 环境科学与生态学
Q1 ENGINEERING, ENVIRONMENTAL
引用次数: 0
Abstract
Due to their chemical inertness, polyethylene (PE) and polystyrene (PS) persistently accumulate in the environment. This study integrates metagenomics, degradation profiling, and molecular simulations to elucidate their divergent microbial degradation pathways. PE degradation was dominated by Burkholderia (97%), with selective C–C bond cleavage causing a 29.8% reduction in weight-average molecular weight (Mw) and a 35.46% degradation rate. PS degradation relied on multispecies cooperation, primarily involving Acinetobacter (52%), Bacillus (21%), and Achromobacter (17%), resulting in random main-chain cleavage, a 9.0% reduction in number-average molecular weight (Mn), and an 18.63% degradation rate. Molecular docking and dynamics simulations showed that PS-degrading enzymes exhibit higher binding affinity (–8.0 kcal/mol) via π–π stacking and cation–π interactions, outperforming the hydrophobic interaction-dominated PE-degrading enzymes (–5.4 kcal/mol). Residue Phe392 exhibited dual functionality in PS degradation for the first time. These findings reveal a divergence in microbial strategies: single-species dominance in PE degradation versus functional consortia for PS. The underlying mechanism is the structural compatibility between polymer substrates and enzyme active sites. This work provides a mechanistic framework for understanding microbial plastic degradation and offers insights for engineering microbial consortia and enzymes for efficient bioremediation of mixed plastic pollution.
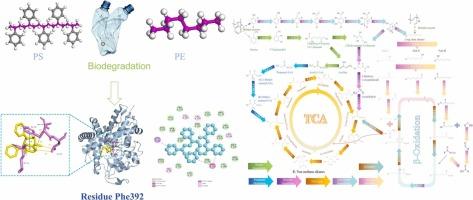
塑料降解的宏基因组和分子模拟:微环境匹配和残留Phe392的关键作用
由于其化学惰性,聚乙烯(PE)和聚苯乙烯(PS)在环境中持续积累。本研究整合宏基因组学、降解谱和分子模拟来阐明它们不同的微生物降解途径。PE的降解以Burkholderia为主(97%),选择性C-C键裂解使PE的分子量(Mw)降低29.8%,降解率降低35.46%。PS的降解依赖于多物种合作,主要涉及不动杆菌(52%)、芽孢杆菌(21%)和无色杆菌(17%),导致主链随机切割,数平均分子量(Mn)降低9.0%,降解率为18.63%。分子对接和动力学模拟表明,ps降解酶通过π -π堆积和阳离子-π相互作用表现出更高的结合亲和力(-8.0 kcal/mol),优于疏水相互作用为主的pe降解酶(-5.4 kcal/mol)。残留Phe392首次在PS降解中表现出双重功能。这些发现揭示了微生物策略的差异:PE降解的单一物种优势与PS的功能联盟。其潜在机制是聚合物底物和酶活性位点之间的结构相容性。这项工作为理解微生物塑料降解提供了一个机制框架,并为有效生物修复混合塑料污染的工程微生物联盟和酶提供了见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Hazardous Materials
工程技术-工程:环境
CiteScore
25.40
自引率
5.90%
发文量
3059
审稿时长
58 days
期刊介绍:
The Journal of Hazardous Materials serves as a global platform for promoting cutting-edge research in the field of Environmental Science and Engineering. Our publication features a wide range of articles, including full-length research papers, review articles, and perspectives, with the aim of enhancing our understanding of the dangers and risks associated with various materials concerning public health and the environment. It is important to note that the term "environmental contaminants" refers specifically to substances that pose hazardous effects through contamination, while excluding those that do not have such impacts on the environment or human health. Moreover, we emphasize the distinction between wastes and hazardous materials in order to provide further clarity on the scope of the journal. We have a keen interest in exploring specific compounds and microbial agents that have adverse effects on the environment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


