Unveiling microbial secondary metabolite biosynthetic gene clusters from alkaline soda Lake Chitu, in the Ethiopian Rift Valley
IF 2.5
4区 生物学
Q3 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Background
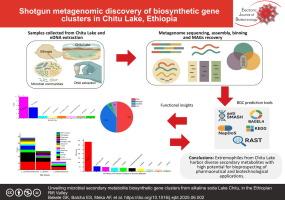
Microorganisms inhabiting alkalihalo-soda lakes are known for producing diverse secondary metabolites with potential biotechnological and pharmaceutical applications. This study explored the biosynthetic capabilities of microbial communities from Ethiopia’s Chitu Lake through shotgun metagenomic sequencing and metagenome-assembled genome (MAG) analyses using various bioinformatics tools.
Results
Analysis of MAGs using the Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) revealed 13 major types of biosynthetic gene clusters. The most abundant were terpene-precursors (32%) and terpene clusters (25%), followed by ribosomally synthesized and post-translationally modified peptides (9%) and nonribosomal peptide synthetases (7%). Other less common BGCs (5% each) included betalactone, ectoine, and Type I polyketide synthase, while rare types (2% each) comprised arylpolyene, hydrogen cyanide, phosphonate, ranthipeptide, and others. The Natural Product Domain Seeker (NaPDoS) detected ketosynthase domains linked to pharmaceutically important such as various fatty acid synthesis, modular and iterative domain classes, and condensation domain which is associated with L-amino acid coupling (LCL) domain class, such as those involved in syringomycin biosynthesis. In addition, bacteriocin analysis identified sactipeptides (56%) and lasso peptides (28%) as dominant types. Kyoto Encyclopedia of Genes and Genomes pathway analysis uncovered several secondary metabolite pathways including those for penicillin, cephalosporins, alkaloids, and phenazines. Rapid Annotation using Subsystem Technology further highlighted secondary metabolism pathways vital for microbial survival in Chitu Lake’s extreme environment.
Conclusions
The discovery of diverse biosynthetic gene cluster positions Chitu Lake as a valuable source of secondary metabolites, highlighting the biotechnological, industrial, pharmaceutical, agricultural and environmental potential of its extremophilic microbes and supporting further bioprospecting efforts.
How to cite: Bekele GK, Balcha ES, Meka AF, et al. Unveiling microbial secondary metabolite biosynthetic gene clusters from alkaline soda Lake Chitu, in the Ethiopian Rift Valley. Electron J Biotechnol 2025;77. https://doi.org/10.1016/j.ejbt.2025.06.002.
揭示来自埃塞俄比亚大裂谷奇图碱性苏打湖的微生物次生代谢物生物合成基因簇
生活在碱盐湖中的微生物以产生多种次生代谢物而闻名,具有潜在的生物技术和制药应用价值。本研究利用各种生物信息学工具,通过散弹枪宏基因组测序和宏基因组组装基因组(MAG)分析,探讨了埃塞俄比亚奇图湖微生物群落的生物合成能力。结果抗菌素和次级代谢物分析软件(antiSMASH)对猪血清进行分析,发现13种主要的生物合成基因簇。最丰富的是萜烯前体(32%)和萜烯簇(25%),其次是核糖体合成和翻译后修饰的肽(9%)和非核糖体肽合成酶(7%)。其他不太常见的bgc(各占5%)包括β内酯、外托因和I型聚酮合成酶,而罕见类型(各占2%)包括芳基多烯、氰化氢、膦酸盐、硫肽等。天然产物结构域搜索器(NaPDoS)检测到与各种重要的药物相关的酮合成酶结构域,如各种脂肪酸合成,模块化和迭代结构域类,以及与l -氨基酸偶联(LCL)结构域类相关的缩合结构域,如紫霉素生物合成。此外,细菌素分析鉴定出sactipeptide(56%)和lasso peptide(28%)为优势类型。京都基因和基因组百科全书通路分析揭示了几种次生代谢物通路,包括青霉素、头孢菌素、生物碱和非那嗪。利用子系统技术的快速注释进一步突出了赤头湖极端环境中微生物生存的次要代谢途径。结论多种生物合成基因簇的发现使赤土湖成为次生代谢产物的宝贵来源,突出了其极端微生物在生物技术、工业、制药、农业和环境方面的潜力,并为进一步的生物勘探工作提供了支持。如何引用:Bekele GK, Balcha ES, Meka AF等。揭示来自埃塞俄比亚大裂谷奇图碱性苏打湖的微生物次生代谢物生物合成基因簇。中国生物医学工程学报(英文版);2009;77。https://doi.org/10.1016/j.ejbt.2025.06.002。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Electronic Journal of Biotechnology
工程技术-生物工程与应用微生物
CiteScore
5.60
自引率
0.00%
发文量
50
审稿时长
2 months
期刊介绍:
Electronic Journal of Biotechnology is an international scientific electronic journal, which publishes papers from all areas related to Biotechnology. It covers from molecular biology and the chemistry of biological processes to aquatic and earth environmental aspects, computational applications, policy and ethical issues directly related to Biotechnology.
The journal provides an effective way to publish research and review articles and short communications, video material, animation sequences and 3D are also accepted to support and enhance articles. The articles will be examined by a scientific committee and anonymous evaluators and published every two months in HTML and PDF formats (January 15th , March 15th, May 15th, July 15th, September 15th, November 15th).
The following areas are covered in the Journal:
• Animal Biotechnology
• Biofilms
• Bioinformatics
• Biomedicine
• Biopolicies of International Cooperation
• Biosafety
• Biotechnology Industry
• Biotechnology of Human Disorders
• Chemical Engineering
• Environmental Biotechnology
• Food Biotechnology
• Marine Biotechnology
• Microbial Biotechnology
• Molecular Biology and Genetics
•Nanobiotechnology
• Omics
• Plant Biotechnology
• Process Biotechnology
• Process Chemistry and Technology
• Tissue Engineering
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


