Capsid structure of phage SPO1 reveals novel minor capsid proteins and insights into capsid stabilization
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
SPO1-related bacteriophages are promising candidates for phage therapy. We present the 3.0 Å cryo-electron microscopy (cryo-EM) structure of the SPO1 capsid with a triangulation number T = 16, enabling the construction of an atomic model comprising the major capsid protein and three types of minor capsid proteins: gp29.2, gp2.7, and gp36.3. These minor capsid proteins adopt novel folds. They might stabilize the capsid and determine its curvature. Gp29.2 monomers contain a three-blade propeller fold and are located at the 3-fold and quasi-three-fold axes. Gp2.7 forms pentamers atop pentameric capsomers, while gp36.3 binds to the capsid’s inner surface, forming star-shaped structures increasing connections between pentameric and hexameric capsomers. The surface exposed regions of gp29.2 and gp2.7 make SPO1 of interest as a nanocage for phage display. Our findings advance the understanding of capsid architecture, stabilization, and local curvature determination for SPO1-related bacteriophages.
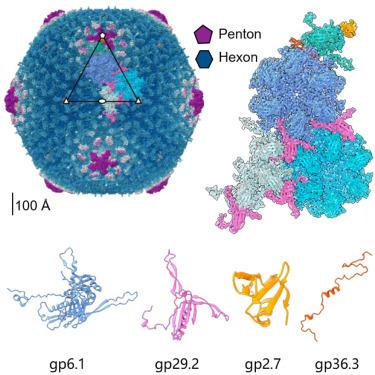
噬菌体SPO1的衣壳结构揭示了新的次要衣壳蛋白和衣壳稳定的见解
spo1相关的噬菌体是噬菌体治疗的有希望的候选者。我们展示了SPO1衣壳的3.0 Å低温电镜(cryo-EM)结构,三角数T = 16,从而构建了一个由主要衣壳蛋白和三种次要衣壳蛋白:gp29.2, gp2.7和gp36.3组成的原子模型。这些次要的衣壳蛋白采用新的折叠。它们可以稳定衣壳并决定其曲率。Gp29.2单体包含一个三叶片螺旋桨折叠,位于三折叠轴和准三折叠轴。Gp2.7在五聚体衣壳体顶部形成五聚体,而gp36.3与衣壳内表面结合,形成星形结构,增加了五聚体和六聚体之间的连接。gp29.2和gp2.7的表面暴露区域使SPO1成为噬菌体展示的纳米笼。我们的发现促进了对spo1相关噬菌体衣壳结构、稳定性和局部曲率测定的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


