A mouse organoid platform for modeling cerebral cortex development and cis-regulatory evolution in vitro
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Natural selection has shaped the gene regulatory networks that orchestrate cortical development, leading to structural and functional variation across mammals, but the molecular and cellular mechanisms underpinning these changes have only begun to be characterized. Here, we develop a reproducible protocol for cerebral cortex organoid generation from mouse epiblast stem cells (EpiSCs), which recapitulates the timing and cellular differentiation programs of the embryonic cortex. We generated cortical organoids from F1 hybrid EpiSCs derived from crosses between laboratory mice (C57BL/6J) and four wild-derived inbred strains spanning ∼1 M years of evolutionary divergence to comprehensively map cis-acting transcriptional regulatory variation across developing cortical cell types, using single-cell RNA sequencing (scRNA-seq). We identify hundreds of genes that exhibit dynamic allelic imbalances, providing the first insight into the developmental mechanisms underpinning changes in cortical structure and function between subspecies. These experimental methods and cellular resources represent a powerful platform for investigating gene regulation in the developing cerebral cortex.
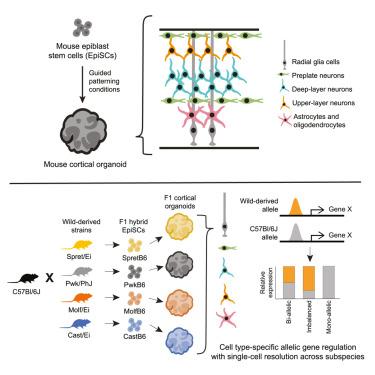
小鼠体外模拟大脑皮层发育和顺式调控进化的类器官平台
自然选择塑造了协调皮质发育的基因调控网络,导致哺乳动物的结构和功能变化,但支撑这些变化的分子和细胞机制才刚刚开始被表征。在这里,我们开发了一个可重复的从小鼠外胚层干细胞(EpiSCs)生成大脑皮层类器官的方案,该方案概括了胚胎皮层的时间和细胞分化程序。我们使用单细胞RNA测序(scRNA-seq)技术,从实验室小鼠(C57BL/6J)与四种跨越1 M年进化差异的野生自交小鼠杂交的F1杂交EpiSCs中生成皮质类器官,以全面绘制发育中的皮质细胞类型的顺式作用转录调控变异。我们发现了数百个表现出动态等位基因失衡的基因,首次深入了解了亚种之间皮层结构和功能变化的发育机制。这些实验方法和细胞资源为研究发育中的大脑皮层的基因调控提供了一个强大的平台。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


