Cross-biobank generalizability and accuracy of electronic health record-based predictors compared to polygenic scores
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
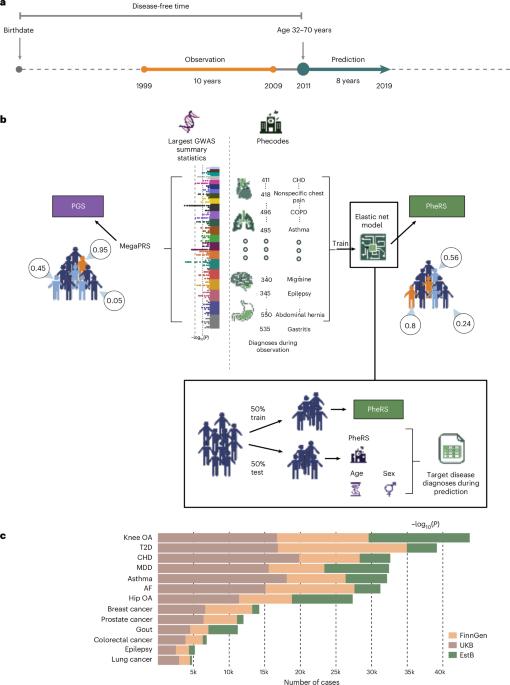
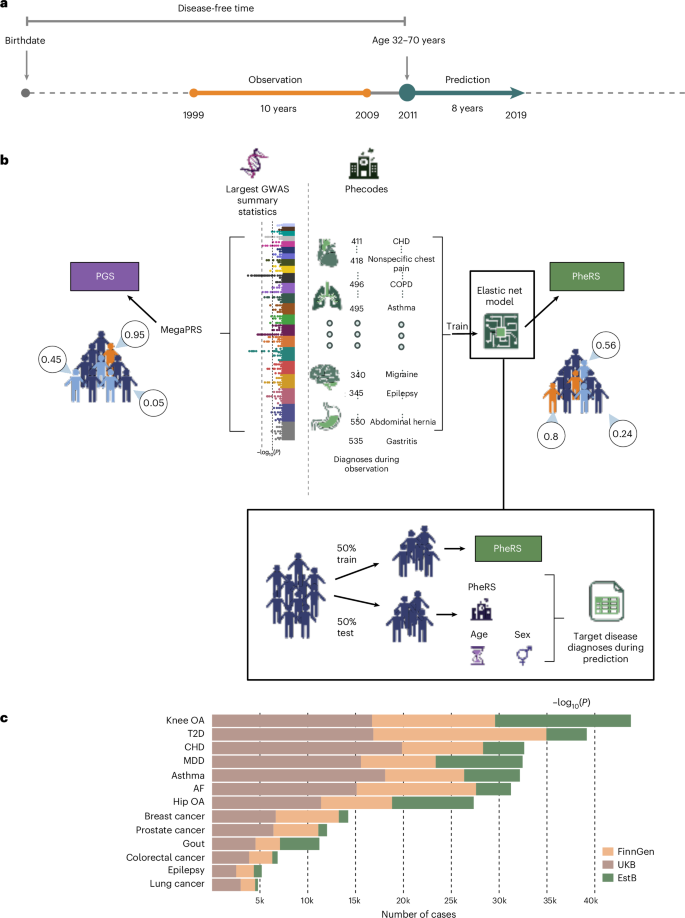
Electronic health record (EHR)-based phenotype risk scores (PheRS) leverage individuals’ health trajectories to estimate disease risk, similar to how polygenic scores (PGS) use genetic information. While PGS generalizability has been studied, less is known about PheRS generalizability across healthcare systems and whether PheRS are complementary to PGS. We trained elastic-net-based PheRS to predict the onset of 13 common diseases for 845,929 individuals (age = 32–70 years) from three biobank-based studies in Finland (FinnGen), the UK (UKB) and Estonia (EstB). All PheRS were statistically significantly associated with the diseases of interest and most generalized well without retraining when applied to other studies. PheRS and PGS were only moderately correlated and models including both predictors improved onset prediction compared to PGS alone for 8 of 13 diseases. Our results indicate that EHR-based risk scores can transfer well between EHRs, capture largely independent information from PGS, and provide additive benefits for disease risk prediction. Comparison of electronic health record-based phenotype risk scores (PheRS) and polygenic scores (PGS) across 13 common diseases and three biobank-based studies indicates that PheRS and PGS may provide additive benefits for risk prediction.
与多基因评分相比,基于电子健康记录的预测指标的跨生物库通用性和准确性
基于电子健康记录(EHR)的表型风险评分(PheRS)利用个体的健康轨迹来估计疾病风险,类似于多基因评分(PGS)使用遗传信息的方式。虽然PGS的普遍性已经被研究过,但关于PheRS在医疗保健系统中的普遍性以及PheRS是否与PGS互补,人们所知甚少。我们训练了基于弹性网络的PheRS来预测来自芬兰(FinnGen)、英国(UKB)和爱沙尼亚(EstB)三个基于生物库的研究的845,929名个体(年龄= 32-70岁)的13种常见疾病的发病。所有PheRS均与相关疾病有统计学显著相关性,并且在应用于其他研究时,大多数患者无需再培训即可很好地普遍化。在13种疾病中,PheRS和PGS仅具有中度相关性,与单独的PGS相比,包含这两种预测因子的模型改善了8种疾病的发病预测。我们的研究结果表明,基于ehr的风险评分可以在ehr之间很好地转移,从PGS中获取很大程度上独立的信息,并为疾病风险预测提供附加效益。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


