Loss of NOTCH2 creates a TRIM28-dependent vulnerability in small cell lung cancer
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Small cell lung cancer (SCLC) is a highly aggressive malignancy that lacks effective targeted therapies, in part due to frequent loss-of-function mutations in tumor suppressors and the absence of recurrent oncogenic drivers. Approximately 15% of SCLCs harbor inactivating mutations in NOTCH1 or NOTCH2, and most neuroendocrine-high SCLCs exhibit low NOTCH activity. Using CRISPR-Cas9 screening in primary cell lines derived from NOTCH1/2-isogenic SCLC genetically engineered mouse models, we identified TRIM28 as a synthetic lethal dependency in NOTCH2-inactivated SCLCs. Loss of TRIM28 in this context robustly induced expression of endogenous retroviruses (ERVs), activated viral sensing pathways, and triggered a type I interferon response. Mechanistically, NOTCH2 inactivation increased reliance on TRIM28-mediated ERV silencing, creating a hyperdependence on TRIM28 via the STING-MAVS-TBK1 axis. Notably, TRIM28 was essential for tumor growth only in the setting of NOTCH2 loss. These findings identify TRIM28 as a potential therapeutic target in NOTCH2-deficient or low-NOTCH2-expressing SCLC.
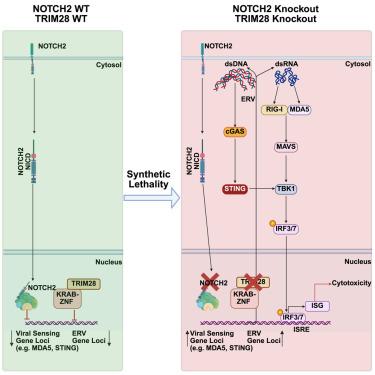
NOTCH2缺失在小细胞肺癌中产生trim28依赖性易感性
小细胞肺癌(SCLC)是一种高度侵袭性的恶性肿瘤,缺乏有效的靶向治疗,部分原因是肿瘤抑制因子中频繁的功能缺失突变和缺乏复发的致癌驱动因素。大约15%的SCLCs携带NOTCH1或NOTCH2失活突变,大多数神经内分泌高的SCLCs表现出低NOTCH活性。利用CRISPR-Cas9筛选来自notch1 /2等基因SCLC基因工程小鼠模型的原代细胞系,我们发现TRIM28在notch2失活的SCLC中是一个合成的致死依赖性。在这种情况下,TRIM28的缺失会强烈诱导内源性逆转录病毒(erv)的表达,激活病毒传感途径,并触发I型干扰素应答。机制上,NOTCH2失活增加了对TRIM28介导的ERV沉默的依赖,通过STING-MAVS-TBK1轴产生对TRIM28的高度依赖。值得注意的是,TRIM28仅在NOTCH2缺失的情况下对肿瘤生长至关重要。这些发现确定TRIM28是notch2缺陷或低notch2表达的SCLC的潜在治疗靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


