Large-scale single-cell profiling of stem cells identifies redundant regulators of shoot development and yield trait variation
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Plant shoot stem cells generate organs essential for food, feed, and biofuels. However, plant single-cell analyses struggled to capture these rare cells or to detect stem cell regulators like CLAVATA3 and WUSCHEL. Here, we dissected stem cell-enriched shoot tissues from maize and Arabidopsis for single-cell RNA sequencing (scRNA-seq), and we optimized protocols to recover thousands of CLAVATA3- and WUSCHEL-expressing cells. A cross-species comparison identified deeply conserved stem cell regulators. We also profiled maize stem cell mutants, validated candidate regulators with spatial transcriptomics, and confirmed their roles in shoot development. These include a family of RNA-binding protein genes and two families of sugar kinase genes. Finally, we show how coupling large-scale single-cell profiling with allelic variation analysis in diverse maize germplasm serves as a valuable resource for identifying stem cell regulators significantly associated with grain yield components. Our discoveries advance the study of shoot stem cells and open avenues for rational crop engineering.
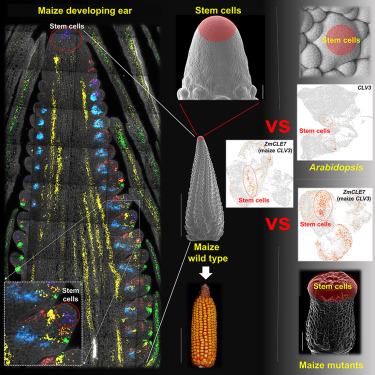
干细胞的大规模单细胞分析鉴定了茎发育和产量性状变异的冗余调节因子
植物芽干细胞产生食物、饲料和生物燃料所必需的器官。然而,植物单细胞分析很难捕获这些罕见的细胞或检测干细胞调节因子,如CLAVATA3和WUSCHEL。在这里,我们解剖了来自玉米和拟南芥的富含干细胞的茎组织进行单细胞RNA测序(scRNA-seq),并优化了方案,以恢复数千个表达CLAVATA3和wuschell的细胞。跨物种比较确定了深度保守的干细胞调节因子。我们还分析了玉米干细胞突变体,用空间转录组学验证了候选调节因子,并确认了它们在茎部发育中的作用。其中包括一个rna结合蛋白基因家族和两个糖激酶基因家族。最后,我们展示了如何将大规模单细胞分析与不同玉米种质的等位基因变异分析相结合,作为鉴定与粮食产量成分显著相关的干细胞调控因子的宝贵资源。我们的发现促进了茎干细胞的研究,为合理的作物工程开辟了道路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


