Uncovering phenotypic inheritance from single cells with Microcolony-seq
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Uncovering phenotypic heterogeneity is fundamental to understanding processes such as development and stress responses. Due to the low mRNA abundance in single bacteria, determining biologically relevant heterogeneity remains a challenge. Using Microcolony-seq, a methodology that captures inherited heterogeneity by analyzing microcolonies originating from single bacterial cells, we uncover the ubiquitous ability of bacteria to maintain long-term inheritance of the host environment. Notably, we observe that growth to stationary phase erases the epigenetic inheritance. By leveraging this memory within each microcolony, Microcolony-seq combines bulk RNA sequencing (RNA-seq) with whole-genome sequencing and phenotypic assays to detect the distinct subpopulations and their fitness advantages. Applying this directly to infected human samples enables us to uncover a wealth of diverse inherited phenotypes. Our observations suggest that bacterial memory may be a widespread phenomenon in both Gram-negative and Gram-positive bacteria. Microcolony-seq provides potential targets for the rational design of therapies with the power to simultaneously target the coexisting subpopulations.
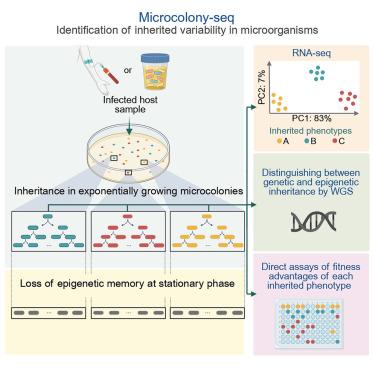
利用Microcolony-seq揭示单细胞的表型遗传
揭示表型异质性是理解发育和应激反应等过程的基础。由于单个细菌的mRNA丰度较低,确定生物学相关的异质性仍然是一个挑战。使用Microcolony-seq(一种通过分析源自单个细菌细胞的微菌落来捕获遗传异质性的方法),我们揭示了细菌普遍存在的维持宿主环境长期遗传的能力。值得注意的是,我们观察到生长到静止阶段消除了表观遗传。通过利用每个微群体的这种记忆,microcolony -seq将大量RNA测序(RNA-seq)与全基因组测序和表型分析相结合,以检测不同的亚群体及其适应度优势。将这种方法直接应用于受感染的人类样本,使我们能够发现丰富多样的遗传表型。我们的观察表明,细菌记忆可能在革兰氏阴性和革兰氏阳性细菌中都是一种普遍现象。Microcolony-seq为合理设计治疗方法提供了潜在的靶标,并具有同时靶向共存亚群的能力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


