Super-pangenome analyses across 35 accessions of 23 Avena species highlight their complex evolutionary history and extensive genomic diversity
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Common oat, belonging to the genus Avena with 30 recognized species, is a nutritionally important cereal crop and high-quality forage worldwide. Here, we construct a genus-level super-pangenome of Avena comprising 35 high-quality genomes from 14 cultivated oat accessions and 21 wild species. The fully resolved phylogenomic analysis unveils the origin and evolutionary scenario of Avena species, and the super-pangenome analysis identifies 26.62% and 59.93% specific genes and haplotypes in wild species. We delineate the landscape of structural variations (SVs) and the transcriptome profile based 1,401 RNA-sequencing (RNA-seq) samples from diverse abiotic stress treatments in oat. We highlight the crucial role of SVs in modulating gene expression and shaping adaptation to diverse stresses. Further combining SV-based genome-wide association studies (GWASs), we characterize 13 candidate genes associated with drought resistance such as AsARF7, validated by transgenic oat lines. Our study provides unprecedented genomic resources to facilitate genomic, evolution and molecular breeding research in oat. A genus-level super-pangenome of Avena comprising 35 high-quality genomes from 23 cultivated or wild oat species highlights the evolution of Avena and the landscape of structural variations related to abiotic stress resistance.

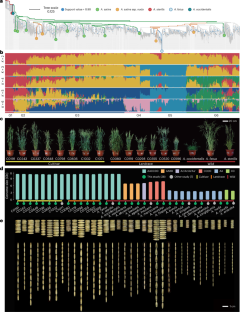
对23个Avena种35份资料的超全基因组分析表明,Avena种具有复杂的进化史和广泛的基因组多样性
普通燕麦属于燕麦属,已知种类30种,是世界范围内重要的营养谷类作物和优质饲料。本研究构建了燕麦属级超泛基因组,包括14个栽培品种和21个野生品种的35个高质量基因组。全解析系统基因组分析揭示了Avena物种的起源和进化情景,超泛基因组分析鉴定出26.62%和59.93%的野生种特异性基因和单倍型。我们描绘了结构变异(SVs)的景观和转录组谱基于1401个rna测序(RNA-seq)样本来自不同的非生物胁迫处理燕麦。我们强调了SVs在调节基因表达和塑造对不同胁迫的适应方面的关键作用。进一步结合基于sv的全基因组关联研究(GWASs),我们鉴定了13个与抗旱性相关的候选基因,如AsARF7,并通过转基因燕麦系验证。我们的研究为燕麦基因组学、进化和分子育种研究提供了前所未有的基因组资源。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


