Microbiome-driven machine learning for predicting suppressiveness to Rhizoctonia solani in organic-amended soils
IF 5
2区 农林科学
Q1 SOIL SCIENCE
引用次数: 0
Abstract
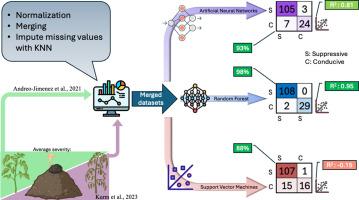
This study introduces a microbiome-integrated machine learning framework to predict soil suppressiveness against Rhizoctonia solani, a destructive fungal pathogen in crops. Combining bacterial and fungal community data, edaphic factors, and disease severity metrics, this study developed robust classification and regression models using Artificial Neural Networks, Random Forest, and Support Vector Machines. Random Forest emerged as the top-performing model, achieving the highest accuracy and explanatory power in classification (98.56 %) and regression (95.40 %) tasks. The study also introduced the Biocontrol Index (BCI), which integrates microbial abundance and predictive importance into interpretable indices linked to soil suppressiveness. Additionally, ecological indices, including alpha diversity, evenness, and beta diversity, were calculated per sample to provide insights into microbial community structure and dynamics. Key predictive features included soil amendments with high microbial activity, carbon-to‑nitrogen ratios, and nutrient levels like calcium and manganese. This integration of microbiome data with machine learning offers a novel, data-driven approach to tackle the high dimensionality of amplicon-sequencing data while also understanding and predicting soil suppressiveness.
微生物组驱动的机器学习预测有机改良土壤对茄枯丝核菌的抑制作用
本研究引入了一个微生物组集成的机器学习框架来预测土壤对作物中破坏性真菌病原体索拉根丝核菌的抑制作用。结合细菌和真菌群落数据、土壤因子和疾病严重程度指标,本研究利用人工神经网络、随机森林和支持向量机建立了稳健的分类和回归模型。随机森林成为表现最好的模型,在分类(98.56%)和回归(95.40%)任务中实现了最高的准确率和解释力。该研究还引入了生物防治指数(BCI),该指数将微生物丰度和预测重要性整合到与土壤抑制相关的可解释指数中。此外,还计算了每个样品的α多样性、均匀度和β多样性等生态指标,以了解微生物群落的结构和动态。关键的预测特征包括具有高微生物活性的土壤改型、碳氮比以及钙和锰等营养水平。这种微生物组数据与机器学习的集成提供了一种新颖的、数据驱动的方法来处理高维的扩增子测序数据,同时也理解和预测土壤的抑制性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Applied Soil Ecology
农林科学-土壤科学
CiteScore
9.70
自引率
4.20%
发文量
363
审稿时长
5.3 months
期刊介绍:
Applied Soil Ecology addresses the role of soil organisms and their interactions in relation to: sustainability and productivity, nutrient cycling and other soil processes, the maintenance of soil functions, the impact of human activities on soil ecosystems and bio(techno)logical control of soil-inhabiting pests, diseases and weeds.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


