Aggresomes protect mRNA under stress in Escherichia coli
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
Membraneless droplets formed through liquid–liquid phase separation of ribonucleoprotein particles contribute to mRNA storage in eukaryotic cells. How such aggresomes contribute to mRNA dynamics under stress and their functional role are less understood in bacteria. Here we used multiple approaches including live-cell imaging, polymer physics modelling and transcriptomics to show that prolonged stress leading to ATP depletion in Escherichia coli results in increased aggresome formation, compaction and selective mRNA enrichment within these aggresomes. Longer transcripts accumulate more in aggresomes than in the cytosol. Mass spectrometry and mutagenesis studies showed that mRNA ribonucleases are excluded from aggresomes due to electrostatic repulsion arising from their negative surface charges. Experiments with fluorescent reporters and disruption of aggresome formation showed that mRNA storage within aggresomes promoted rapid translation reactivation and is associated with reduced lag phases during growth after stress removal. Our findings suggest that mRNA storage within aggresomes confers an advantage for bacterial survival and recovery from stress. Ribonucleoprotein aggresomes exclude ribonucleases and protect mRNA to promote rapid translation reactivation and cellular recovery after stress alleviation in Escherichia coli.
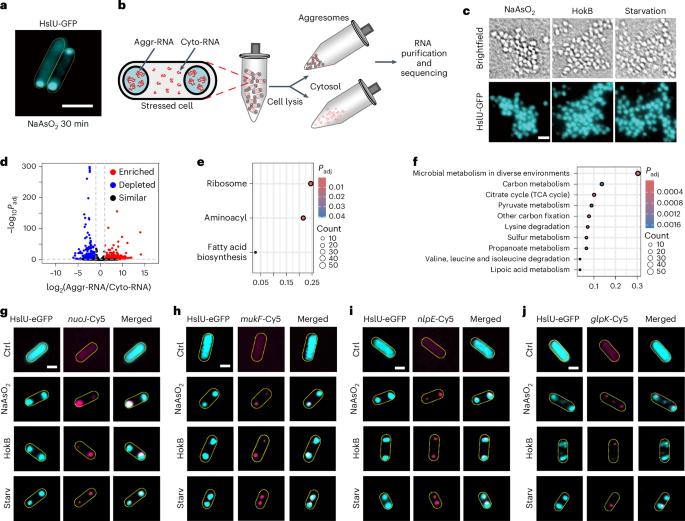
大肠杆菌中聚集体在应激条件下保护mRNA。
核糖核蛋白颗粒通过液-液相分离形成的无膜液滴有助于真核细胞中mRNA的储存。在细菌中,这些聚合体如何在压力下促进mRNA动态及其功能作用尚不清楚。在这里,我们使用了多种方法,包括活细胞成像、聚合物物理建模和转录组学来证明,长期应激导致大肠杆菌中ATP的消耗,导致这些聚合体形成、压实和选择性mRNA富集增加。较长的转录本在聚合体中比在细胞质中积累更多。质谱和诱变研究表明,mRNA核糖核酸酶由于其表面负电荷引起的静电斥力而被排除在聚合体之外。荧光报告和破坏聚合体形成的实验表明,聚合体内的mRNA储存促进了快速翻译再激活,并与去除胁迫后生长过程中的滞后期减少有关。我们的研究结果表明,聚合体中的mRNA储存对细菌的生存和从压力中恢复具有优势。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


