Systematic decoding of functional enhancer connectomes and risk variants in human glioma
IF 19.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Genetic and epigenetic variations contribute to the progression of glioma, but the mechanisms underlying these effects, particularly for enhancer-associated genetic variations in non-coding regions, still remain unclear. Here we performed high-throughput CRISPR interference screening to identify pro-tumour enhancers in glioma cells. By integrating genome-wide H3K27ac HiChIP data, we identified the target genes of these pro-tumour enhancers and revealed the essential role of enhancer connectomes in promoting glioma progression. Through systematic analysis of enhancers carrying glioma risk-associated single-nucleotide polymorphisms (SNPs), we found that these SNPs can promote glioma progression through the enhancer connectome. Using CRISPR–Cas9-mediated enhancer interference and SNP editing, we demonstrated that glioma-specific enhancer carrying the risk SNP rs2297440 regulates SOX18 expression by specifically recruiting transcription factor MEIS1 binding, thereby contributing to glioma progression. Our study sheds light on the molecular mechanisms underlying glioma susceptibility and provides potential therapeutic targets to treat glioma. Bi, Mo, Liu et al. carry out high-throughput screening and analysis to profile genome-wide pro-tumour enhancers and uncover enhancer connectomes in glioma. In particular, they identify specific single-nucleotide polymorphisms associated with glioma risk and progression.

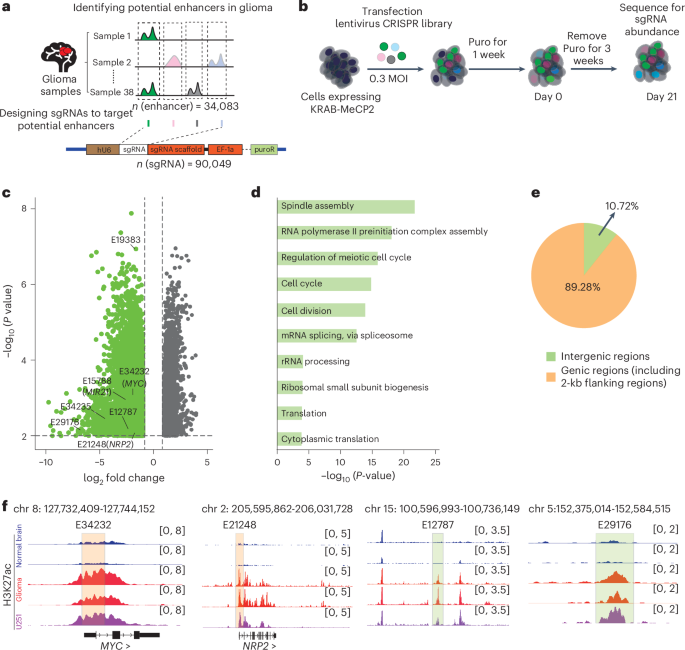
人类胶质瘤中功能增强子连接体和风险变异的系统解码。
遗传和表观遗传变异有助于胶质瘤的进展,但这些影响的机制,特别是在非编码区增强子相关的遗传变异,仍然不清楚。在这里,我们进行了高通量CRISPR干扰筛选,以鉴定胶质瘤细胞中的促肿瘤增强子。通过整合全基因组H3K27ac HiChIP数据,我们确定了这些促肿瘤增强子的靶基因,并揭示了增强子连接体在促进胶质瘤进展中的重要作用。通过系统分析携带胶质瘤风险相关单核苷酸多态性(SNPs)的增强子,我们发现这些SNPs可以通过增强子连接组促进胶质瘤的进展。通过crispr - cas9介导的增强子干扰和SNP编辑,我们证明了携带风险SNP rs2297440的胶质瘤特异性增强子通过特异性募集转录因子MEIS1结合来调节SOX18的表达,从而促进胶质瘤的进展。我们的研究揭示了胶质瘤易感性的分子机制,并为治疗胶质瘤提供了潜在的治疗靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


